#install.packages("ggtern")
#install.packages("patchwork")
#install.packages("tidypaleo")3-Gráficos com ggplot2
Aula 3
Dr. Gabriel Bertolini
2º semestre 2023 - PPGGEO UFRGS
Oficina de programação.
Aula introdutória sobre gráficos com ggplot.
Instalar pacotes
Importar biblioteca
library(dplyr) # biblioteca do tidyverse de manipulação de banco de dados
library(tidyr) # biblioteca do tidyverse para transformar banco de dados
library(ggtern) # biblioteca que adiciona o gráfico ternário ao ggplot (evitar importar ggplot2 ao importar ggtern)
library(forcats) # biblioteca do tidyverse para lidar com fatores
library(patchwork) # biblioteca para organizar gráficos em um layout
library(tidypaleo) # biblioteca para plots estratigráficos
library(RColorBrewer) # biblioteca de paleta de cores
library(gt)
# Bibliotecas espaciais
library(sf) # biblioteca para criação de objetos espaciais
library(mapview) # biblioteca para visualização interativa de dados espaciais
library(geobr)
library(ggspatial)
library(ggdal)Importar os bancos
Banco obtido em @bonin2020 .
BD<-read.csv("./data/geoquimica_granito.csv") %>%
as_tibble()
BD %>%
gt_preview()| X.1 | X | Tectonic_setting | Location_notes | Pluton | Colour | Symbol | Size_rel | Size | SubGroup | Group | DebonPQ | TASMiddlemostPlut | Villaseca | QANOR | LaRoche | Geol_unit_notes | Rock_type_notes | Ref | Age | Latitude | Longitude | SiO2 | TiO2 | Al2O3 | FeOt | MnO | MgO | CaO | K2O | Na2O | P2O5 | H2O | H2Ot | H2O.MINUS | LOI | Li | Be | B | Sc | V | Cr | Ni | Cu | Zn | Rb | Sr | Y | Zr | Nb | Cs | Ba | La | Ce | Pr | Nd | Sm | Eu | Gd | Tb | Dy | Ho | Er | Tm | Yb | Lu | Hf | Ta | Pb | Th | U | Co | Mo | W | Ga | Ge | As | In | Sn | Sb | Cd | Q | C | Or | Ab | An | Ne | Ns | Ks | Di | Wo | Hy | Ol | Il | Tn | Pf | Ru | Ap | Sum_CIPW | Orthoclase | Albite | Anorthite | Quartz | Apatite | Ilmenite | Biotite | Amphibole | Corundum | Rest | sum_meso | P | K | Ti | Cr2O3 | A.NK | A.CNK | K2O.Na2O | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | CR03 | OROGENIC BELT | ////////Peninsula////// | Cape Peninsula | brown4 | 16 | 1.6 | 1.28 | Mafic S-types | 1- CPG & MPG | gr | quartz monzonite | h-P | 3b granite | granodiorite | ////////// | ////// | Unpub. Data, A.Villaros, G. Stevens, R. Scheepers (Villaros et al. 2009) | NA | NA | 65.40690 | 0.8270369 | 17.08490 | 4.914410 | 0.07714897 | 1.7960280 | 1.813515 | 5.131949 | 2.660096 | 0.2880228 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 269.0154 | 97.21508 | 52.24591 | 305.2263 | 25.35902 | NA | 390.4759 | 56.40085 | 112.16620 | 13.223848 | 52.34722 | 11.393739 | 1.2494124 | 10.838077 | 1.6265380 | 9.835326 | 2.024970 | 6.206267 | 0.8976863 | 6.077240 | NA | 8.747229 | NA | NA | 26.782157 | NA | NA | NA | NA | NA | NA | 24.54893 | 4.546013 | 30.32835 | 22.50899 | 7.115483 | 0 | 0 | 0 | 0 | 0 | 4.474137 | 0 | 0.16503878 | 0 | 0 | 0.7405025 | 0.6822185 | 95.10966 | 25.62016 | 22.53101 | 7.109292 | 27.20246 | 0.6797338 | 0.7856851 | 6.932014 | 0 | 4.544585 | -0.3193515 | 95.08560 | 1257.1043 | 42601.85 | 4958.086 | NA | 1.720334 | 1.291518 | 1.929235 | ||||||||
| 2 | 2 | BB202 | OROGENIC BELT | ////////Peninsula////// | Cape Peninsula | brown4 | 16 | 1.6 | 1.28 | Mafic S-types | 1- CPG & MPG | gr | granodiorite | h-P | 3b granite | granodiorite | ////////// | ////// | Unpub. Data, A.Villaros, G. Stevens, R. Scheepers (Villaros et al. 2009) | NA | NA | 69.64232 | 0.7087655 | 14.88713 | 4.186309 | 0.05397209 | 1.6293460 | 1.436880 | 5.018386 | 2.215911 | 0.22098 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 214.2795 | 98.96108 | 38.28057 | 260.1457 | 19.91177 | NA | 415.6909 | 44.29357 | 90.02860 | 10.382656 | 39.39793 | 7.655565 | 1.0286014 | 8.160105 | 1.1358404 | 8.071263 | 1.516150 | 3.946216 | 0.5438978 | 3.740191 | NA | 7.342101 | NA | NA | 19.492634 | NA | NA | NA | NA | NA | NA | 32.66955 | 3.726173 | 29.65722 | 18.75042 | 5.684928 | 0 | 0 | 0 | 0 | 0 | 4.058911 | 0 | 0.11545828 | 0 | 0 | 0.6482846 | 0.5234192 | 95.83437 | 25.43081 | 18.76877 | 5.680122 | 35.07460 | 0.5215128 | 0.6733272 | 6.222967 | 0 | 3.726782 | -0.2852007 | 95.81369 | 964.4893 | 41659.13 | 4249.049 | NA | 1.640003 | 1.273484 | 2.264706 | ||||||||
| 3 | 3 | CB02 | OROGENIC BELT | ////////Peninsula////// | Cape Peninsula | brown4 | 16 | 1.6 | 1.28 | Mafic S-types | 1- CPG & MPG | gr | granite | h-P | 3b granite | granite | ////////// | ////// | Unpub. Data, A.Villaros, G. Stevens, R. Scheepers (Villaros et al. 2009) | NA | NA | 70.61077 | 0.5705854 | 15.28393 | 2.822104 | 0.05318077 | 1.3306273 | 1.204323 | 5.597277 | 2.329983 | 0.197212 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 238.5302 | 90.38265 | 29.96433 | 186.0415 | 15.13822 | NA | 547.5218 | 32.36167 | 63.93163 | 7.248746 | 28.7628 | 6.040634 | 1.076092 | 5.3337 | 0.8584825 | 5.574644 | 1.088590 | 3.174243 | 0.4446103 | 2.755472 | NA | 5.434977 | NA | NA | 14.798949 | NA | NA | NA | NA | NA | NA | 31.63569 | 3.674628 | 33.07830 | 19.71567 | 4.686449 | 0 | 0 | 0 | 0 | 0 | 3.314764 | 0 | 0.11376549 | 0 | 0 | 0.5109352 | 0.4671216 | 97.19733 | 29.62021 | 19.73496 | 4.682256 | 33.59253 | 0.4654203 | 0.5420561 | 5.098139 | 0 | 3.676313 | -0.2339899 | 97.17789 | 860.7515 | 46464.68 | 3420.659 | NA | 1.545114 | 1.265067 | 2.402282 | ||||||||
| 4 | 4 | HB02 | OROGENIC BELT | ////////Peninsula////// | Cape Peninsula | brown4 | 16 | 1.6 | 1.28 | Mafic S-types | 1- CPG & MPG | gr | granite | h-P | 3b granite | granite | ////////// | ////// | Unpub. Data, A.Villaros, G. Stevens, R. Scheepers (Villaros et al. 2009) | NA | NA | 70.61724 | 0.4775028 | 15.70859 | 2.508109 | 0.04066072 | 0.9779424 | 1.311569 | 5.280680 | 2.877528 | 0.2001758 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 254.2501 | 116.29881 | 36.86868 | 181.9237 | 15.15780 | NA | 499.1298 | 34.12151 | 70.41246 | 8.06835 | 30.55275 | 6.613892 | 1.2179917 | 6.696204 | 1.0960613 | 7.270367 | 1.316706 | 3.774652 | 0.5311750 | 3.354008 | NA | 5.152241 | NA | NA | 15.483984 | NA | NA | NA | NA | NA | NA | 29.97338 | 3.353324 | 31.20731 | 24.34884 | 5.199155 | 0 | 0 | 0 | 0 | 0 | 2.436180 | 0 | 0.08698231 | 0 | 0 | 0.4319138 | 0.4741418 | 97.51123 | 28.63039 | 24.37266 | 5.194790 | 31.40372 | 0.4724149 | 0.4536277 | 3.789478 | 0 | 3.351382 | -0.1765739 | 97.49189 | 873.6873 | 43836.51 | 2862.629 | NA | 1.503237 | 1.223927 | 1.835145 | ||||||||
| 5 | 5 | CR01 | OROGENIC BELT | ////////Peninsula////// | Cape Peninsula | brown4 | 16 | 1.6 | 1.28 | Mafic S-types | 1- CPG & MPG | gr | granite | h-P | 3a granite | granite | ////////// | ////// | Unpub. Data, A.Villaros, G. Stevens, R. Scheepers (Villaros et al. 2009) | NA | NA | 70.70878 | 0.5322106 | 15.28031 | 2.535746 | 0.06085569 | 1.2071555 | 1.235924 | 5.569955 | 2.642243 | 0.2268257 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 233.7445 | 91.32626 | 38.96812 | 172.6349 | 14.63283 | NA | 408.2444 | 32.54389 | 62.92283 | 7.49205 | 29.71011 | 6.455299 | 1.0194147 | 6.264685 | 1.0442407 | 6.880438 | 1.272111 | 3.816830 | 0.4902676 | 3.014657 | NA | 4.650457 | NA | NA | 14.926382 | NA | NA | NA | NA | NA | NA | 30.22193 | 3.200335 | 32.91684 | 22.35792 | 4.649750 | 0 | 0 | 0 | 0 | 0 | 3.007180 | 0 | 0.13018383 | 0 | 0 | 0.4638989 | 0.5372654 | 97.48530 | 29.75506 | 22.37980 | 4.645108 | 31.99386 | 0.5353087 | 0.5056001 | 4.664697 | 0 | 3.200603 | -0.2157710 | 97.46426 | 990.0035 | 46237.87 | 3190.603 | NA | 1.472673 | 1.210504 | 2.108040 | ||||||||
| 6..3049 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3050 | 3050 | SGMV | OROGENIC BELT | Carajas Province, Amazonia | Velho Guilherme Suite | lightgreen | 18 | 2.0 | 1.60 | PostOro (A2) | 5- KCG | Teixeira et al. 2019 | 1860-1880 | NA | NA | 75.95000 | 0.0600000 | 12.62000 | 2.745275 | NA | 0.0100000 | 0.580000 | 4.530000 | 2.590000 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 786.0000 | 18.00000 | 170.00000 | 164.0000 | 76.00000 | NA | 28.0000 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
Banco obtido em @bertolini2020
lvsd<-read.csv("./data/lvsd.csv") %>%
as_tibble()
lvsd %>%
gt_preview()| X | sample | well | depth | h1 | h2 | h3 | h4 | h5 | h6 | h7 | h8 | h9 | h10 | h_mean | pw_t1 | pw_t2 | pw_t3 | sw_t1 | sw_t2 | p_wave | s_wave | dry_mass | subm_mass | sat_mass | bulk_vol | pore_volum | porosity | density | lava_1 | lava_2 | lava_dist_base | exp_1 | exp_2 | total_points | comp_quartz | comp_feldspar | comp_lithic | qtz_mono | qtz_poly | qtz_deformed | f_plagio | f_microcline | f_ortoclase | lt_metamorphic | lt_plutonic | lt_vulcanic | lt_sedimentary | por_primary | por_moldic | por_intragranular | por_total | por_IGV | con_float | con_point | con_linear | con_cc | con_sutured | con_tontal | cem_zeolite | cem_qtz | cem_calcite | cem_semctite | cem_ilite | cem_albite | cem_chloriet | cem_ironoxide | cem_gypsum | cem_dolomite | cem_total | gs_silt | gs_vfs | gs_fs | gs_ms | gs_cs | gs_vcs | gs_sorting | gm_high_sphericity | gm_low_sphericity | gm_angular | gm_subangular | gm_subrounded | gm_rounded | lava_1_base | lava_1_top | lava_2_base | lava_2_top | dis_1_top | dis_1_base | dis_2_top | dis_2_base | dis_l1 | dis_l2 | distance | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | RB-01 | TG-27 | 1.25 | 25.98 | 26.86 | 25.70 | 26.56 | 25.46 | 25.93 | 26.62 | 25.28 | 26.45 | 26.48 | 26.132 | 4.78 | 4.68 | NA | 7.38 | 7.33 | 5.52 | 3.55 | 50.45 | 29.35 | 50.50 | 21.15 | 0.05 | 0.24 | 2.39 | 97.0 | NA | 89.7 | 7.4 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 97.0 | 89.6 | NA | NA | 88.35 | -95.75 | NA | NA | 88.35 | NA | 88.35 |
| 2 | 2 | RB-02 | TG-27 | 1.90 | 44.92 | 44.93 | 45.04 | 44.94 | 44.78 | 44.80 | 44.88 | 44.92 | 45.03 | 44.89 | 44.913 | 13.63 | 13.93 | NA | 18.63 | 18.83 | 3.26 | 2.40 | 52.10 | 31.81 | 55.33 | 23.52 | 3.23 | 13.73 | 2.22 | 97.0 | NA | 89.7 | 7.4 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 97.0 | 89.6 | NA | NA | 87.70 | -95.10 | NA | NA | 87.70 | NA | 87.70 |
| 3 | 3 | RB-03 | TG-27 | 2.90 | 44.04 | 43.49 | 43.50 | 42.96 | 42.76 | 43.45 | 43.58 | 43.44 | 43.56 | 43.71 | 43.449 | 13.53 | 12.73 | NA | 15.93 | 15.63 | 3.31 | 2.75 | 35.26 | 26.50 | 37.76 | 11.26 | 2.50 | 22.20 | 3.13 | 97.0 | NA | 89.7 | 7.4 | NA | 356 | 64.47368 | 21.71053 | 13.815789 | 93.36735 | 0.5102041 | 6.122449 | 43.54839 | 53.22581 | 3.225806 | 5.128205 | 76.92308 | 15.384615 | 2.564103 | 16.01124 | 0.2808989 | 4.775281 | 21.06742 | 23.59551 | 0.2139037 | 28.44920 | 42.8877 | 21.28342 | 7.165775 | 100 | 0 | 0 | 2 | 5 | 0 | 0 | 0 | 2 | 0 | 0 | 9 | 4.109589 | 8.904110 | 21.91781 | 28.08219 | 33.56164 | 3.424658 | 1.204 | 53.62319 | 46.37681 | 24.087591 | 18.24818 | 39.41606 | 18.24818 | 97.0 | 89.6 | NA | NA | 86.70 | -94.10 | NA | NA | 86.70 | NA | 86.70 |
| 4 | 4 | RB-04 | TG-27 | 3.90 | 47.45 | 46.67 | 46.45 | 46.14 | 46.52 | 47.06 | 47.43 | 48.79 | 47.82 | 47.68 | 47.201 | 13.68 | 13.73 | NA | 16.23 | 16.93 | 3.44 | 2.85 | 54.56 | 34.03 | 58.86 | 24.83 | 4.30 | 17.32 | 2.20 | 97.0 | NA | 89.7 | 7.4 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 97.0 | 89.6 | NA | NA | 85.70 | -93.10 | NA | NA | 85.70 | NA | 85.70 |
| 5 | 5 | RB-05 | TG-27 | 4.90 | 45.59 | 45.43 | 45.28 | 45.34 | 45.30 | 45.24 | 45.20 | 45.25 | 45.28 | 45.36 | 45.327 | 13.48 | 12.58 | NA | 14.23 | 14.23 | 3.48 | 3.19 | 56.21 | 35.50 | 59.39 | 23.89 | 3.18 | 13.31 | 2.35 | 97.0 | NA | 89.7 | 7.4 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 97.0 | 89.6 | NA | NA | 84.70 | -92.10 | NA | NA | 84.70 | NA | 84.70 |
| 6..71 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 72 | 72 | RB-71 | CBM-100UF | 95.00 | 34.55 | 34.44 | 34.79 | 35.01 | 35.80 | 36.32 | 36.57 | 35.89 | 35.20 | 34.77 | 35.334 | 8.48 | 8.83 | NA | 11.03 | 12.63 | 4.08 | 2.99 | 131.68 | 77.95 | 138.79 | 60.84 | 7.11 | 11.69 | 2.16 | 4.7 | 89.7 | 5.9 | 4.8 | 85 | 312 | 67.61566 | 23.84342 | 8.540925 | 95.78947 | 0.0000000 | 4.210526 | 33.92857 | 51.78571 | 14.285714 | 16.666667 | 75.00000 | 8.333333 | 0.000000 | 10.89744 | 0.3205128 | 1.602564 | 12.82051 | 17.62821 | 0.0000000 | 4.61361 | 52.7105 | 34.25606 | 8.419839 | 100 | 3 | 0 | 4 | 0 | 0 | 7 | 0 | 0 | 1 | 0 | 15 | 1.204819 | 2.409639 | 16.86747 | 32.53012 | 45.78313 | 1.204819 | 0.915 | 70.19868 | 29.80132 | 0.621118 | 22.36025 | 56.52174 | 20.49689 | 4.7 | -0.1 | 89.7 | 4.7 | -95.10 | 90.30 | -90.3 | 5.3 | 90.30 | 5.3 | 5.30 |
Banco dummy de paleocorrents
PALEOCURRENTS<-read.csv("paleocurrents_dummy.csv")
PALEOCURRENTS %>%
gt_preview()| X | direction | facies | |
|---|---|---|---|
| 1 | 1 | 53.06491 | WINDRIPPLES |
| 2 | 2 | 40.29703 | WINDRIPPLES |
| 3 | 3 | 73.98197 | WINDRIPPLES |
| 4 | 4 | 67.22886 | WINDRIPPLES |
| 5 | 5 | 66.95277 | WINDRIPPLES |
| 6..899 | |||
| 900 | 900 | 149.29671 | DUNES |
Gráficos ggplot2
Scatterplot
PLOT1<-BD %>%
select(SiO2,TiO2) %>%
ggplot(aes(x=TiO2,y=SiO2))+
#parametro aes()=aestetic para parametro
#relacionado ao banco de dado (coluna)
geom_point(size=2)+
#tipo de elemento gráfico a ser usado,
#no caso pontos
scale_x_log10()
#parametro adicionado para usar
#escala log10 no eixo x
PLOT1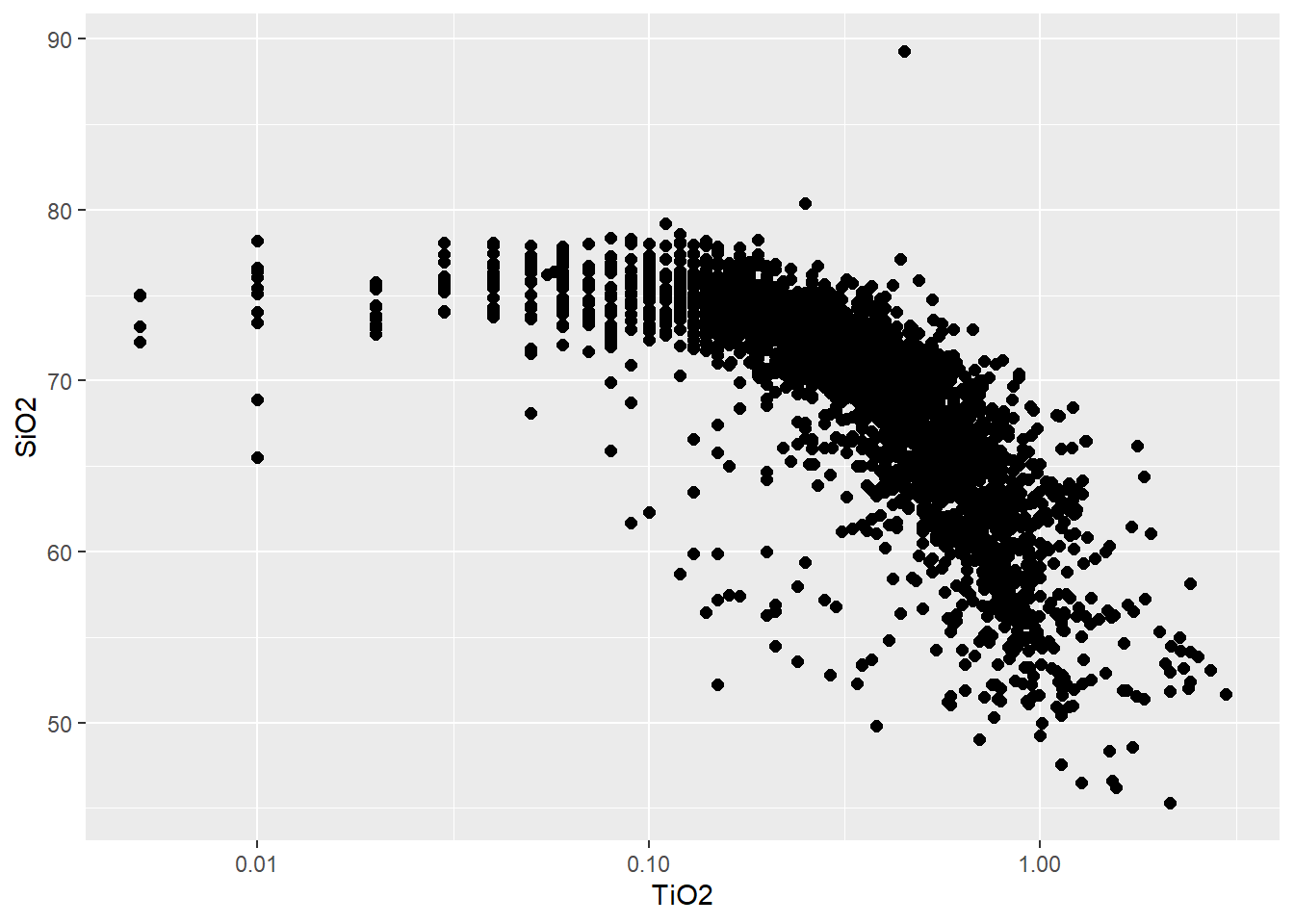
Gráfico interativo com o plotly
plotly::ggplotly(PLOT1)Aqui, vamos fazer um gráfico de dispersão utilizando diferentes transparencias, adicionadas com função mutate e criação da coluna transp. Notar uso do aes() para mapear o banco de dados.
BD %>%
select(SiO2,TiO2,MgO,Tectonic_setting) %>%
mutate(transp=ifelse(Tectonic_setting=="CONTINENTAL FLOOD BASALT",0.8,0.3)) %>%
ggplot(aes(x=SiO2,y=TiO2,size=MgO,color=Tectonic_setting))+
#geom_point(alpha=0.5) # Parametro alpha para transparência
geom_point(aes(alpha=transp))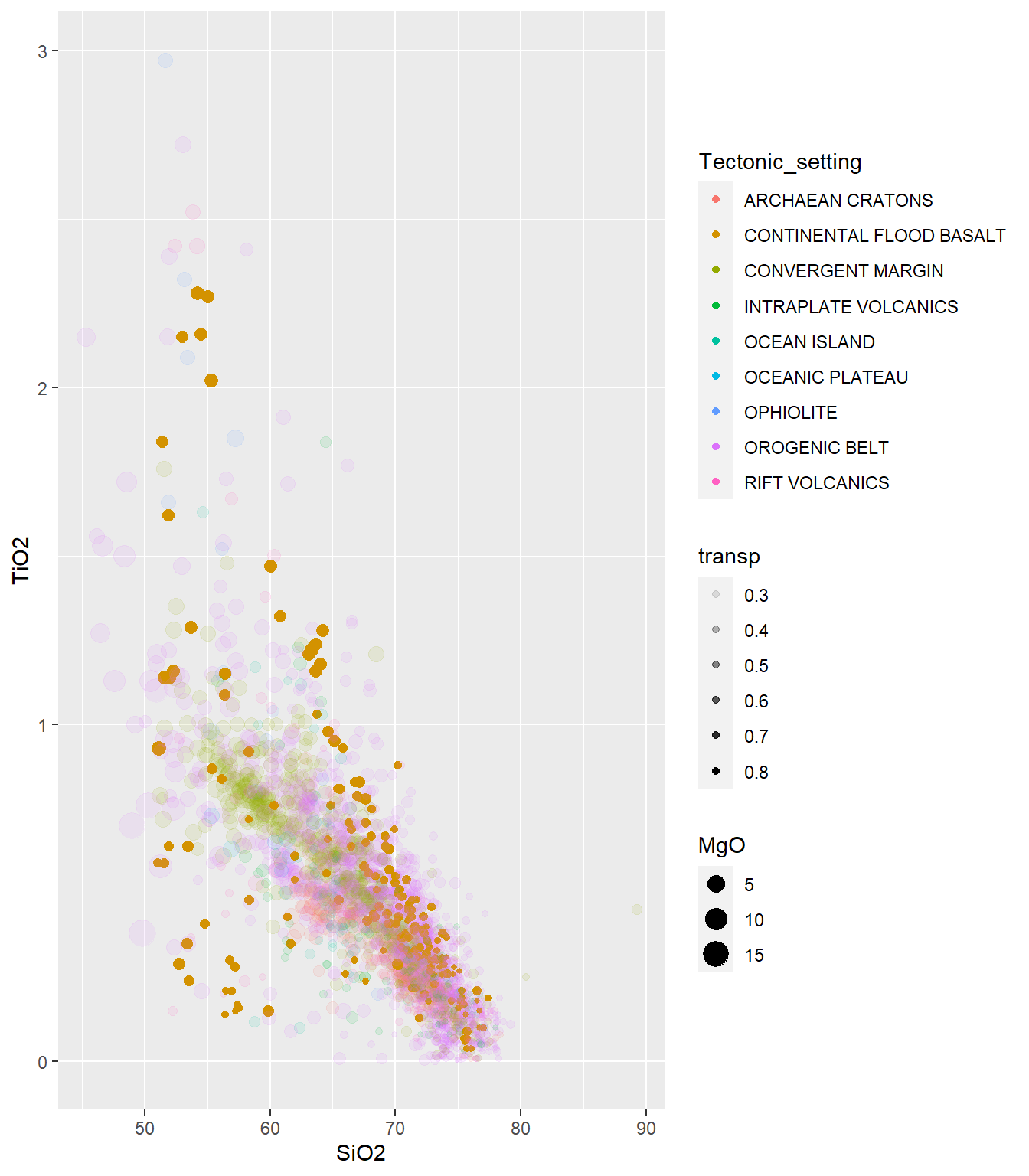
A função ggsave() pode salvar o plot.
#ggsave("./plots/plot1.jpg")Plot múltiplos com facet_wrap()
BD %>%
select(SiO2,TiO2,MgO,Tectonic_setting) %>%
ggplot(aes(x=SiO2,y=TiO2,size=MgO,col=Tectonic_setting))+
geom_point()+
labs(x="Óxido de Silicio",
y="Óxido de Titânio",
size="Proporção de Magnésio",
col="Ambiente tectonico",
title = " Relação entre Silicio e Titânio em diferentes ambientes geotectônicos")+
facet_wrap(~Tectonic_setting)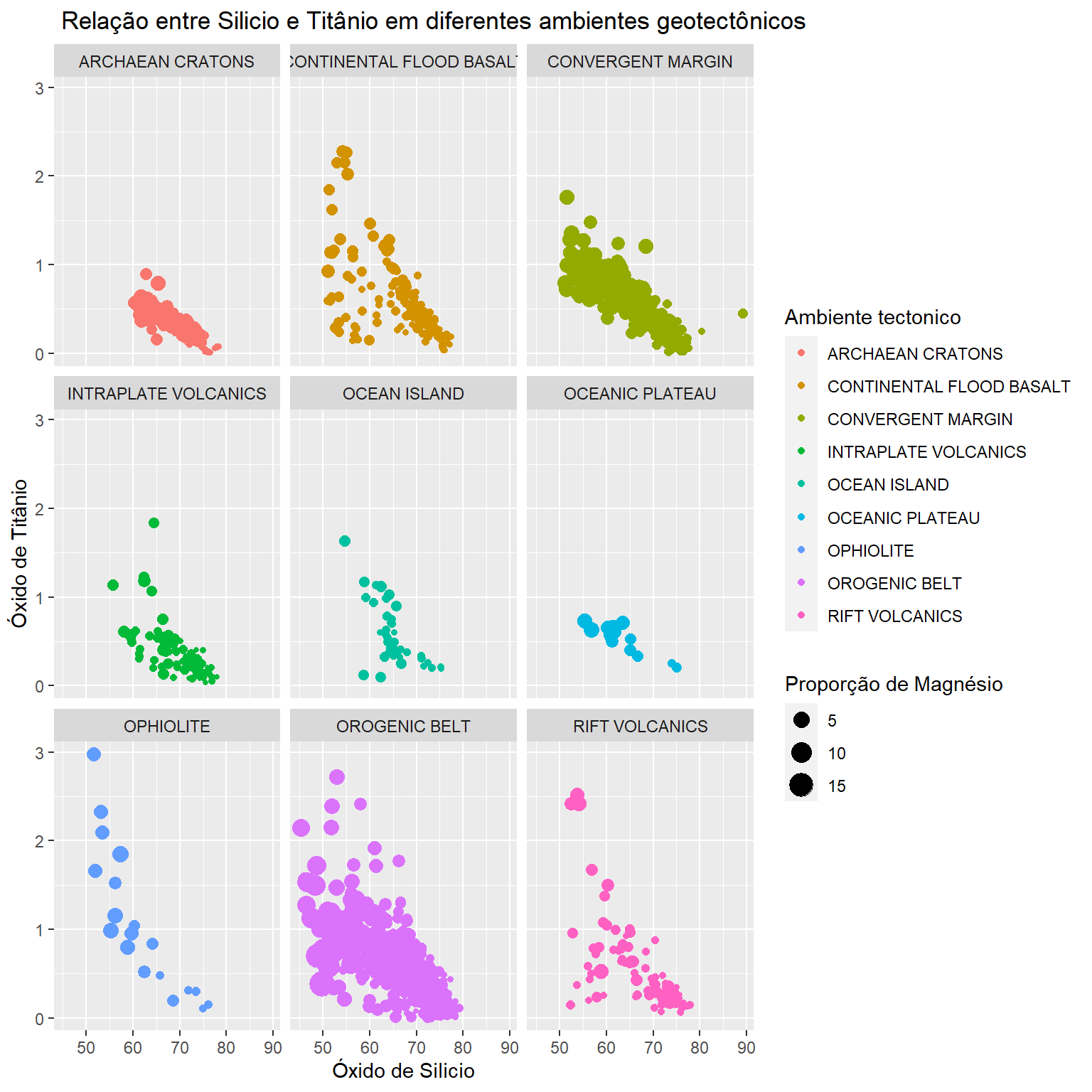
ou facet_grid()
BD %>%
select(SiO2,TiO2,MgO,Tectonic_setting,TASMiddlemostPlut,SubGroup) %>%
filter(Tectonic_setting=="ARCHAEAN CRATONS") %>%
ggplot(aes(x=SiO2,y=TiO2,col=SubGroup))+
geom_point()+
facet_grid(TASMiddlemostPlut~SubGroup)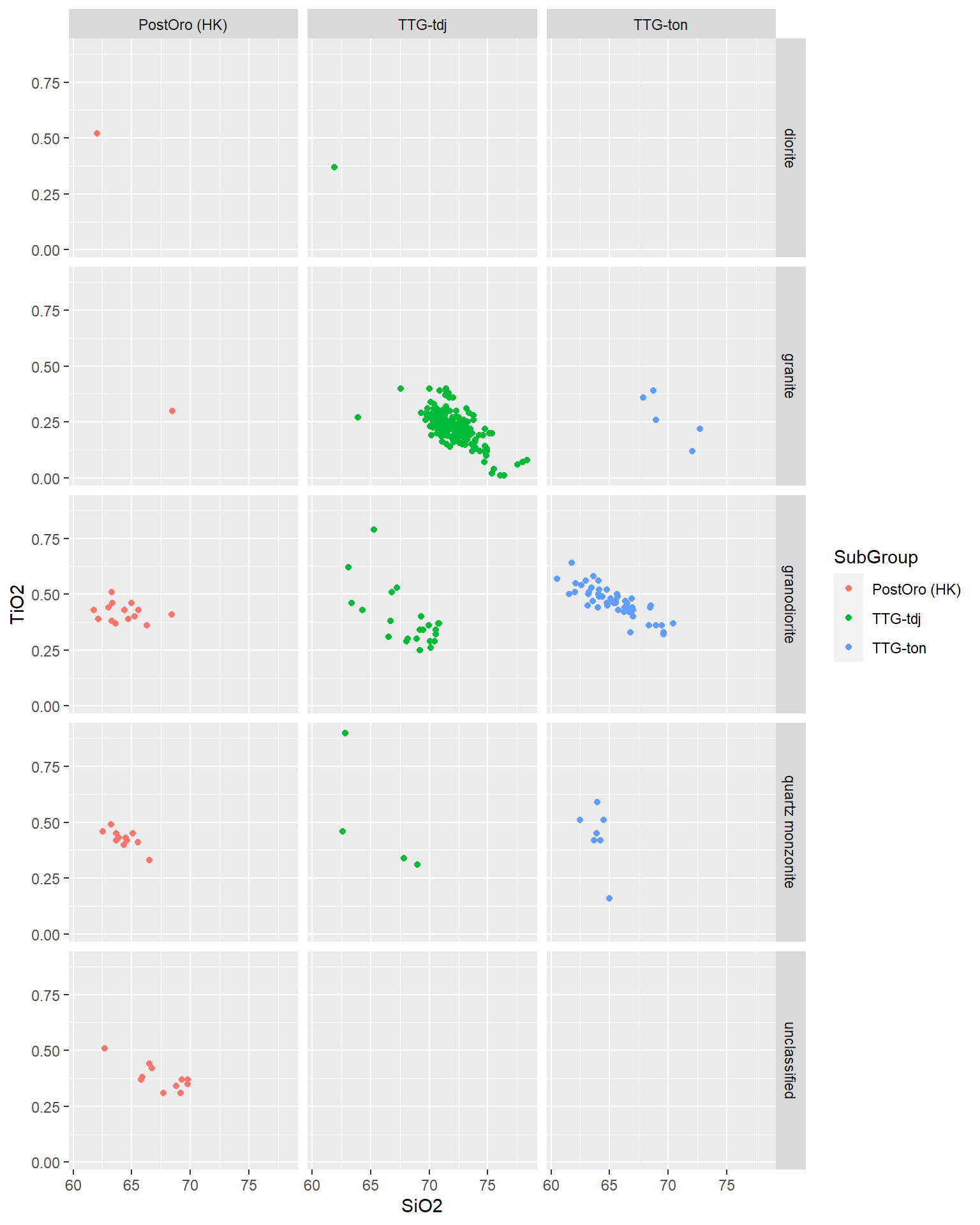
Exercício 1
Qual a distribuição das séries de granitos utilizando o diagrama P-Q (Debon & Fort, 1983, 1988)
\[ P = K - (Na + Ca) \]
\[ Q = \frac{Si}{3} - (K + Na + 2 \times \frac{Ca}{3}) \]
P vs Q
- Fazer gráfico de dispersão P vs. Q no banco dos granitos
BD %>% select(K=K2O,Na=Na2O,Si=SiO2,Ca=CaO,SubGroup) %>%
mutate(P = K - (Na + Ca),
Q = Si/3 - (K + Na + 2*(Ca/3))) %>%
ggplot(aes(P,Q,col=SubGroup))+
geom_point()+
geom_smooth(method="lm")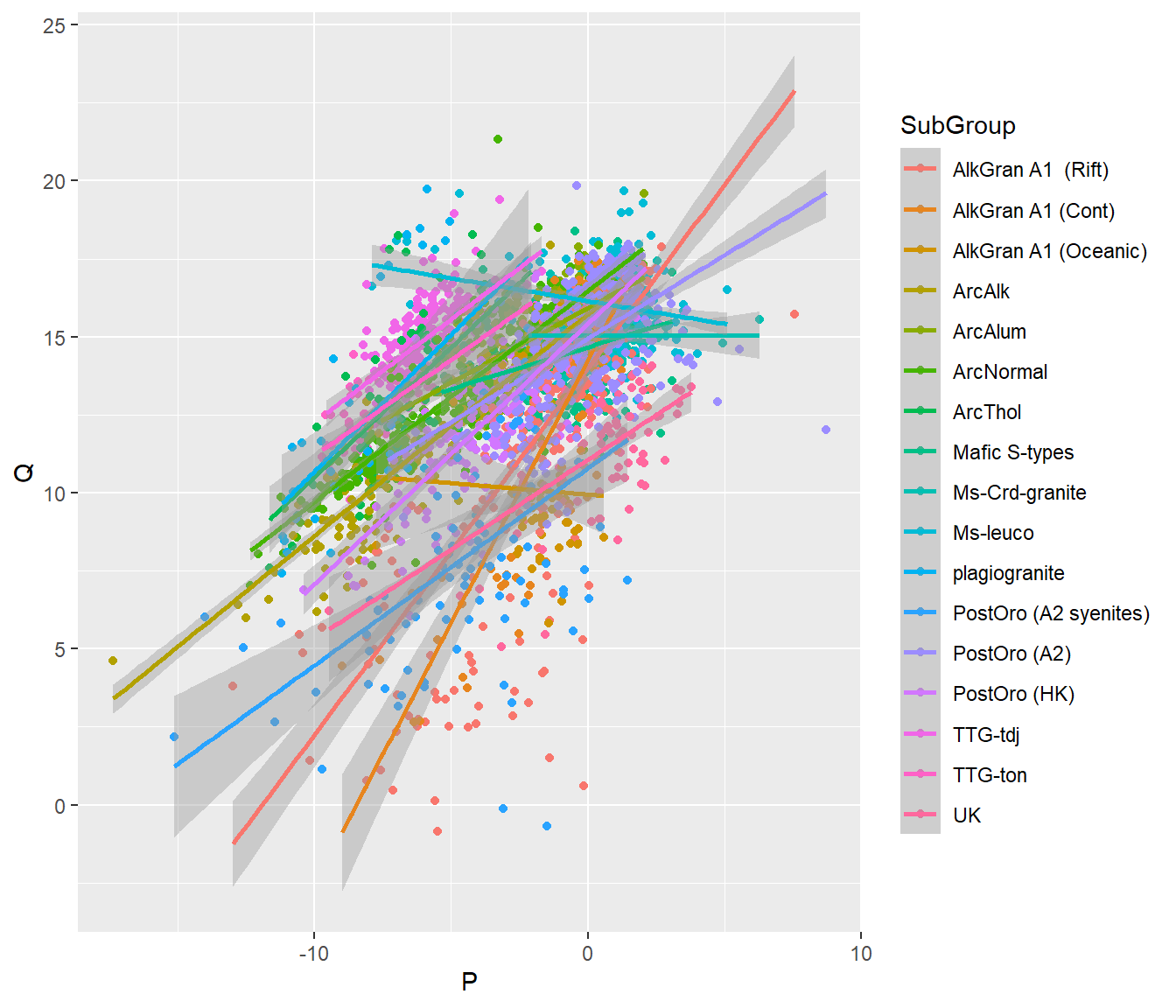
- Criar vários plots pela coluna SubGroup.
BD %>% select(K=K2O,Na=Na2O,Si=SiO2,Ca=CaO,SubGroup) %>%
mutate(P = K - (Na + Ca),
Q = Si/3 - (K + Na + 2*(Ca/3))) %>%
ggplot(aes(P,Q,col=SubGroup))+
geom_point()+
geom_smooth(method="lm",col="black")+
facet_wrap(~SubGroup)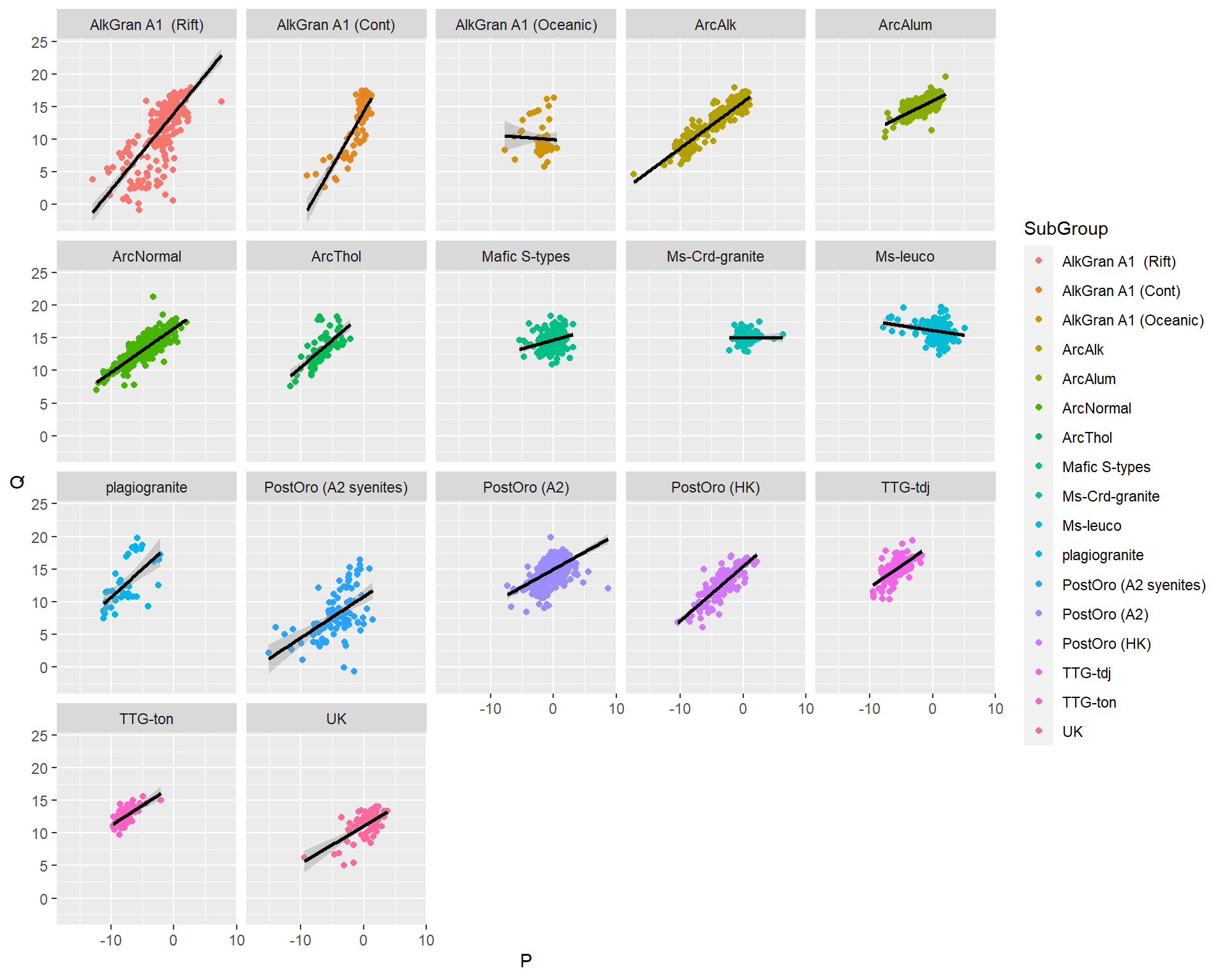
Boxplot
BD %>%
select(SiO2, TiO2, MgO, Tectonic_setting) %>%
ggplot(aes(x = Tectonic_setting, y = TiO2, fill = Tectonic_setting)) +
geom_boxplot()+
theme(axis.title.x=element_blank(), # retirar nome escala
axis.text.x=element_blank(), # retirar texto escala
axis.ticks.x=element_blank()) # retirar ticks escala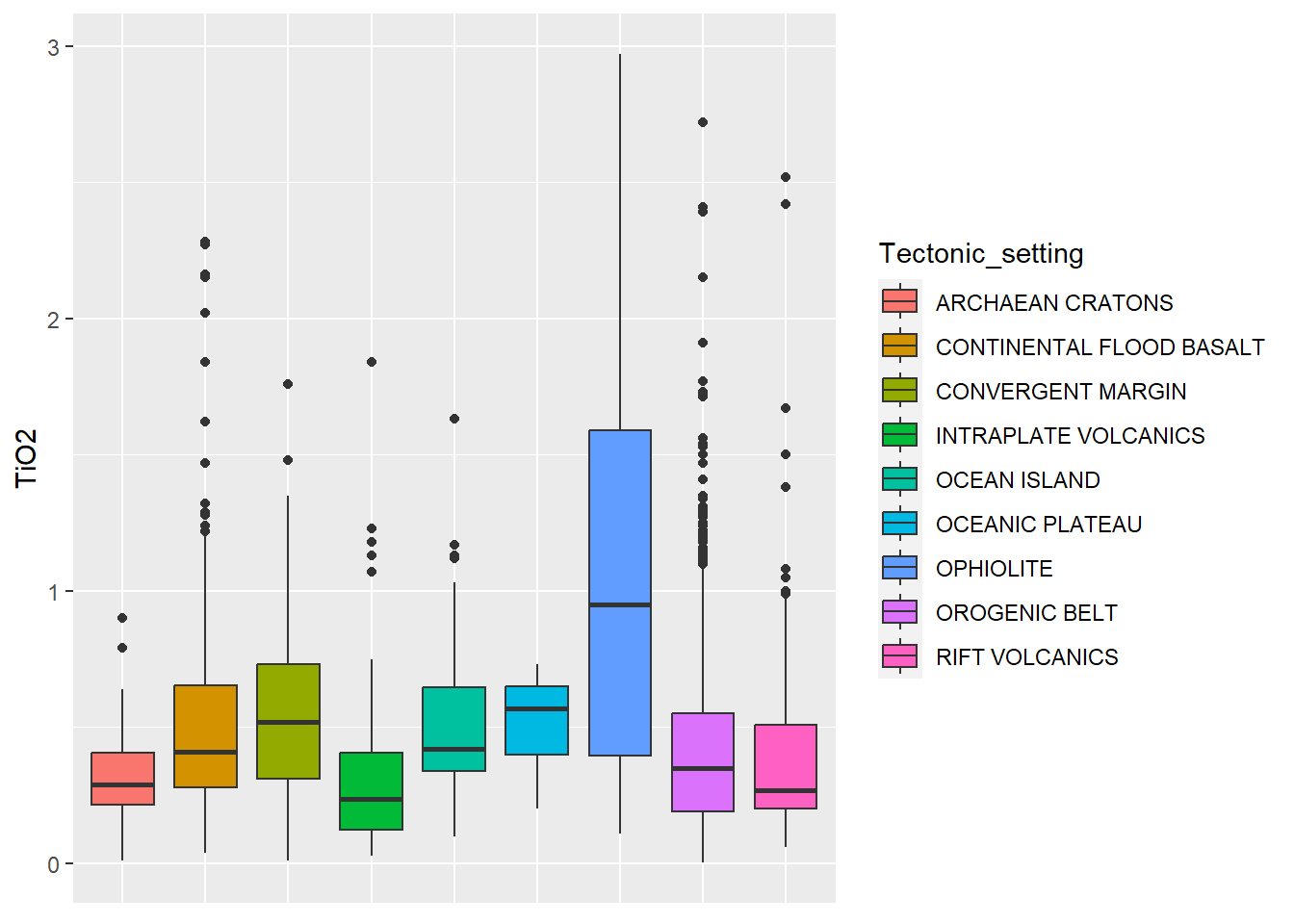
Ou, versão violino
BD %>%
select(SiO2, TiO2, MgO, Tectonic_setting) %>%
ggplot(aes(x = Tectonic_setting, y = TiO2, fill = Tectonic_setting)) +
geom_violin()+
theme(axis.title.x=element_blank(), # retirar nome escala
axis.text.x=element_blank(), # retirar texto escala
axis.ticks.x=element_blank()) # retirar ticks escala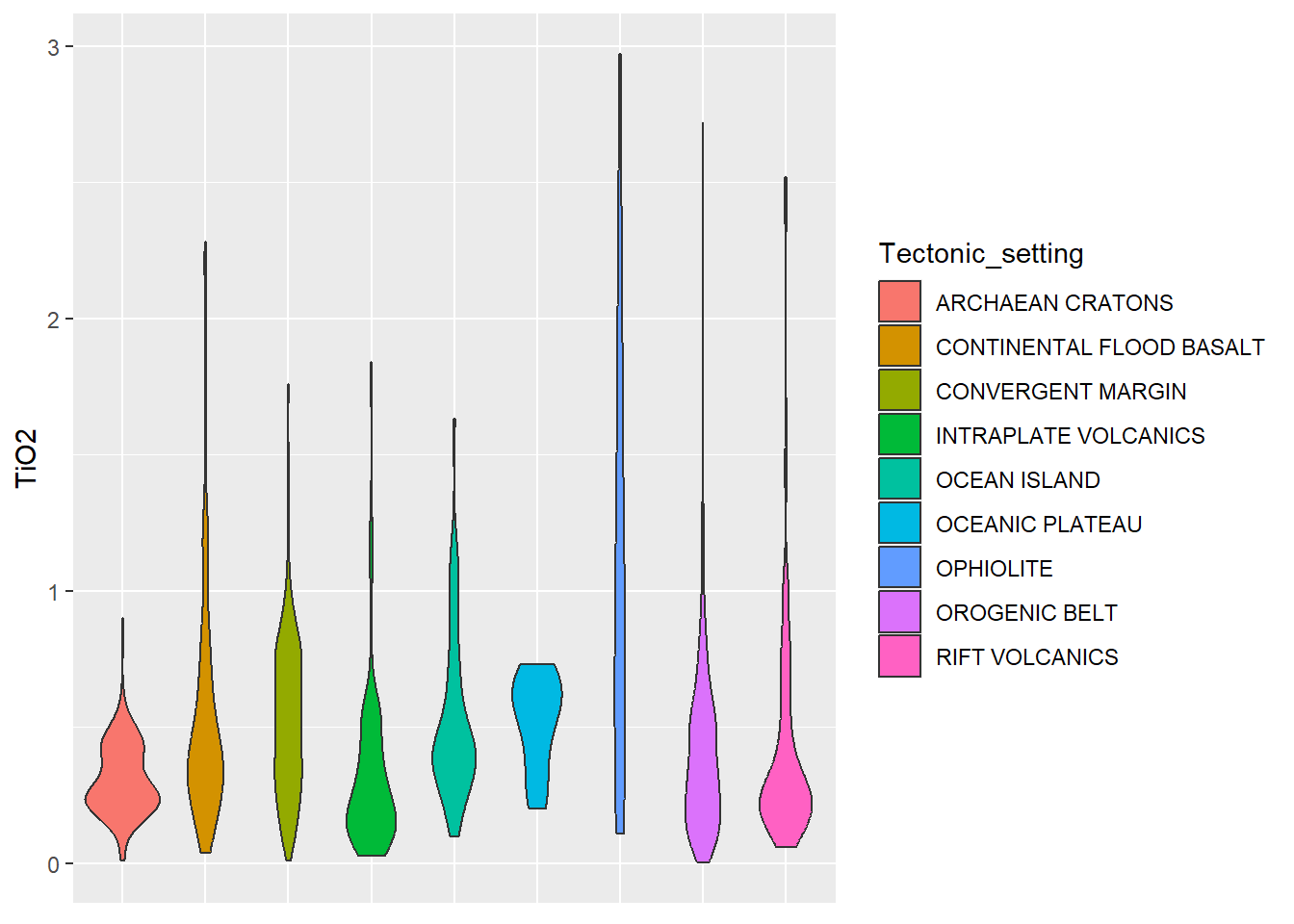
Histograma
BD %>% select(SiO2,TiO2,MgO,Tectonic_setting) %>%
ggplot(aes(x=SiO2))+
geom_histogram(bins=10)+
geom_vline(aes(xintercept=mean(SiO2)))+ # Adiciona média
geom_vline(col="red",aes(xintercept=median(SiO2))) # adiciona mediana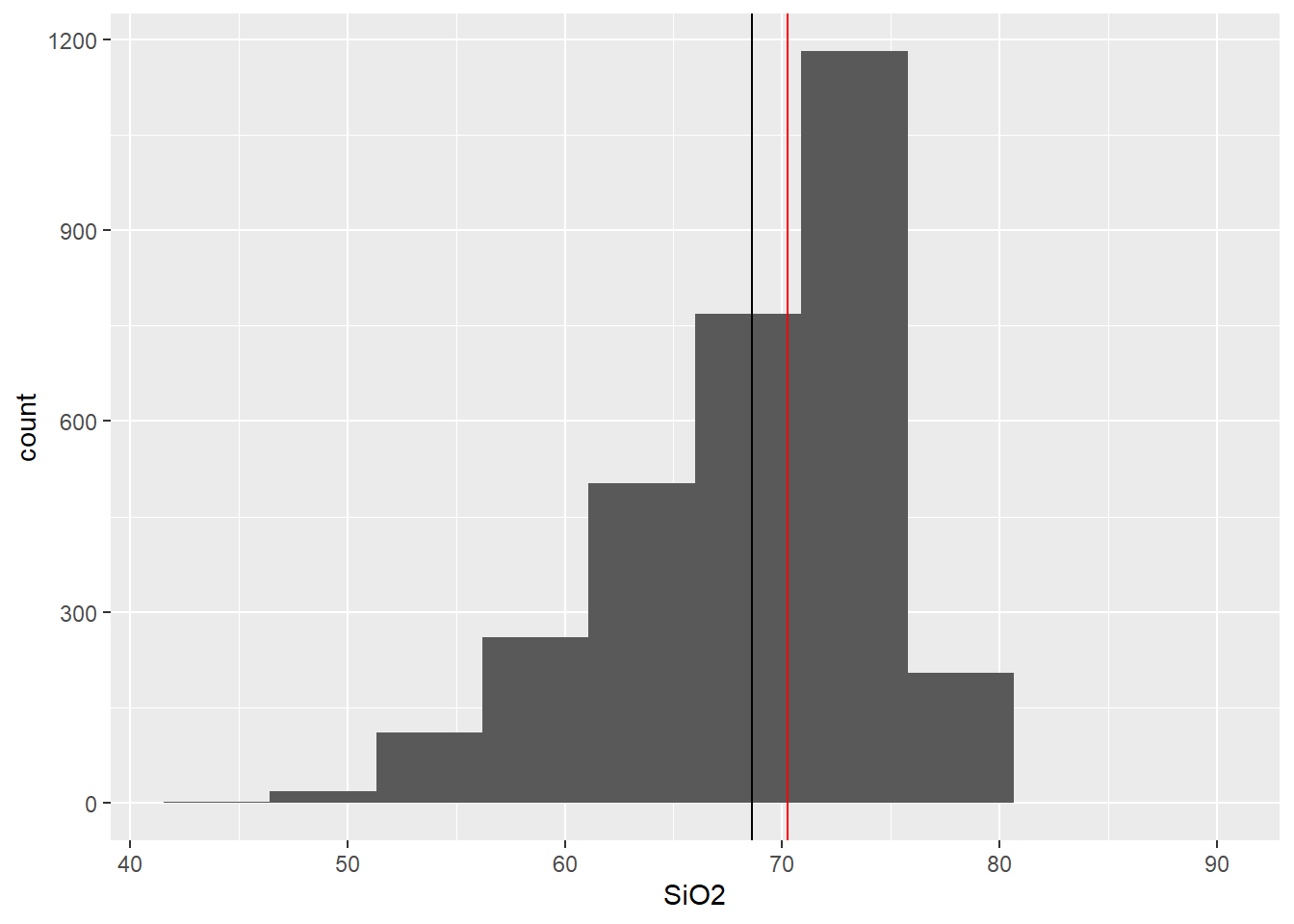
histo_ploter <- function(df,bins = 10) {
df %>%
ggplot(aes(x = SiO2) ) +
geom_histogram(bins = bins) +
geom_vline(aes(xintercept = mean(SiO2))) +
geom_vline(col = "red", aes(xintercept = median(SiO2)))
}
histo_ploter(BD,5)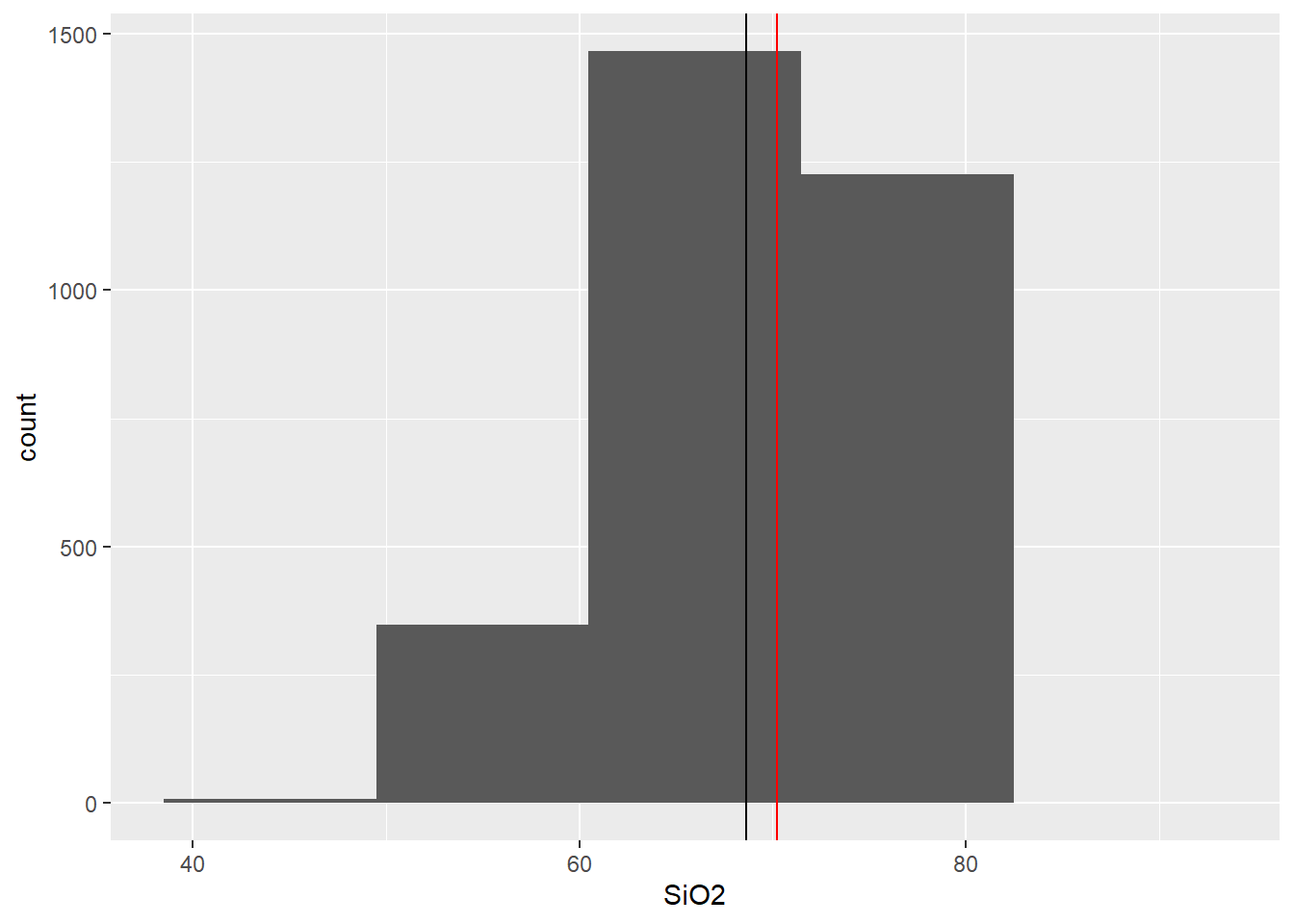
plotly::ggplotly(histo_ploter(BD))Histograma pivotado
BD %>%
select(SiO2:P2O5) %>%
mutate(across(.cols = SiO2:P2O5, as.numeric)) %>%
pivot_longer(cols = SiO2:P2O5,
names_to = "element",
values_to = "values") %>%
group_by(element) %>%
mutate(media = median(values, na.rm = TRUE)) %>%
ungroup() %>%
ggplot(aes(values)) +
geom_histogram() +
geom_vline(col = "red", aes(xintercept = media)) +
facet_wrap( ~ element, scales = "free")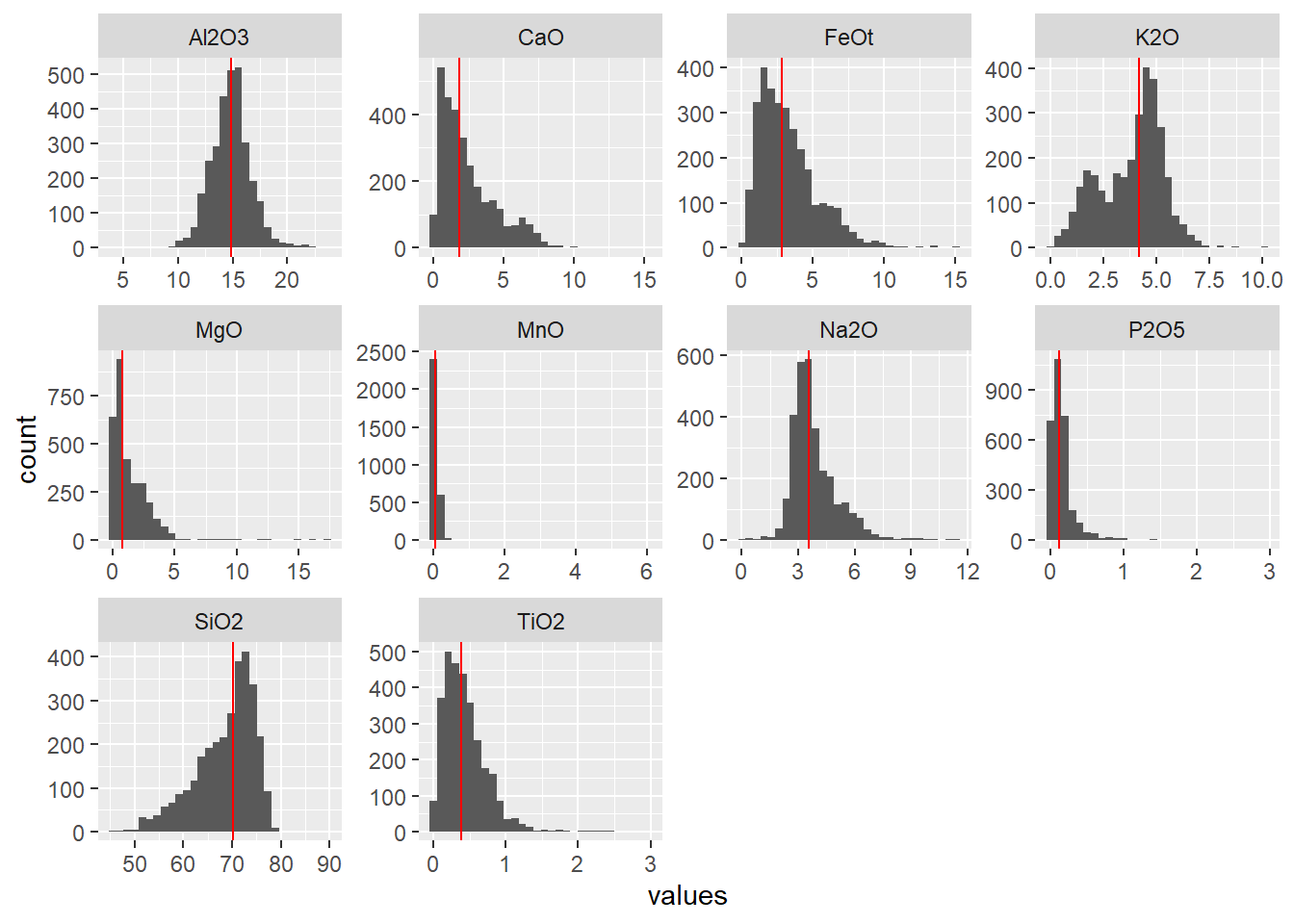
Densidade
BD %>%
select(SiO2, Tectonic_setting) %>%
ggplot(aes(x = SiO2, fill = Tectonic_setting)) +
geom_density(bw=1, #tamanho da banda
alpha = 0.6) +
facet_wrap( ~ Tectonic_setting)+
labs(fill="Ambiente\nTectônico")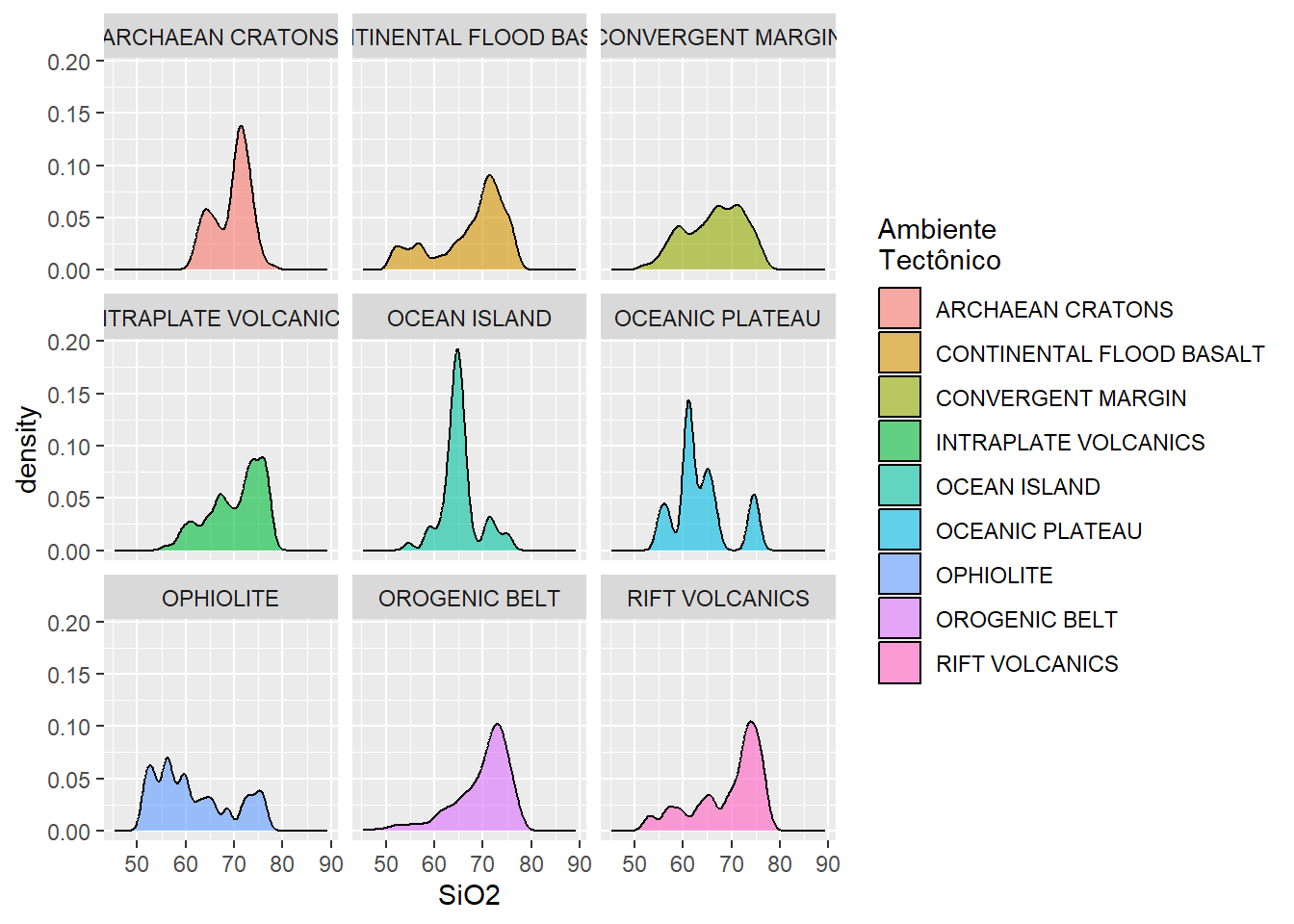
Ternário
simples
BD %>%
filter(Tectonic_setting=="OROGENIC BELT") %>%
select(Q = Quartz,
Ab = Albite,
An = Anorthite) %>%
ggtern(aes(x=Ab, y=Q, z=An)) +
geom_point() +
ggtern::geom_confidence_tern() 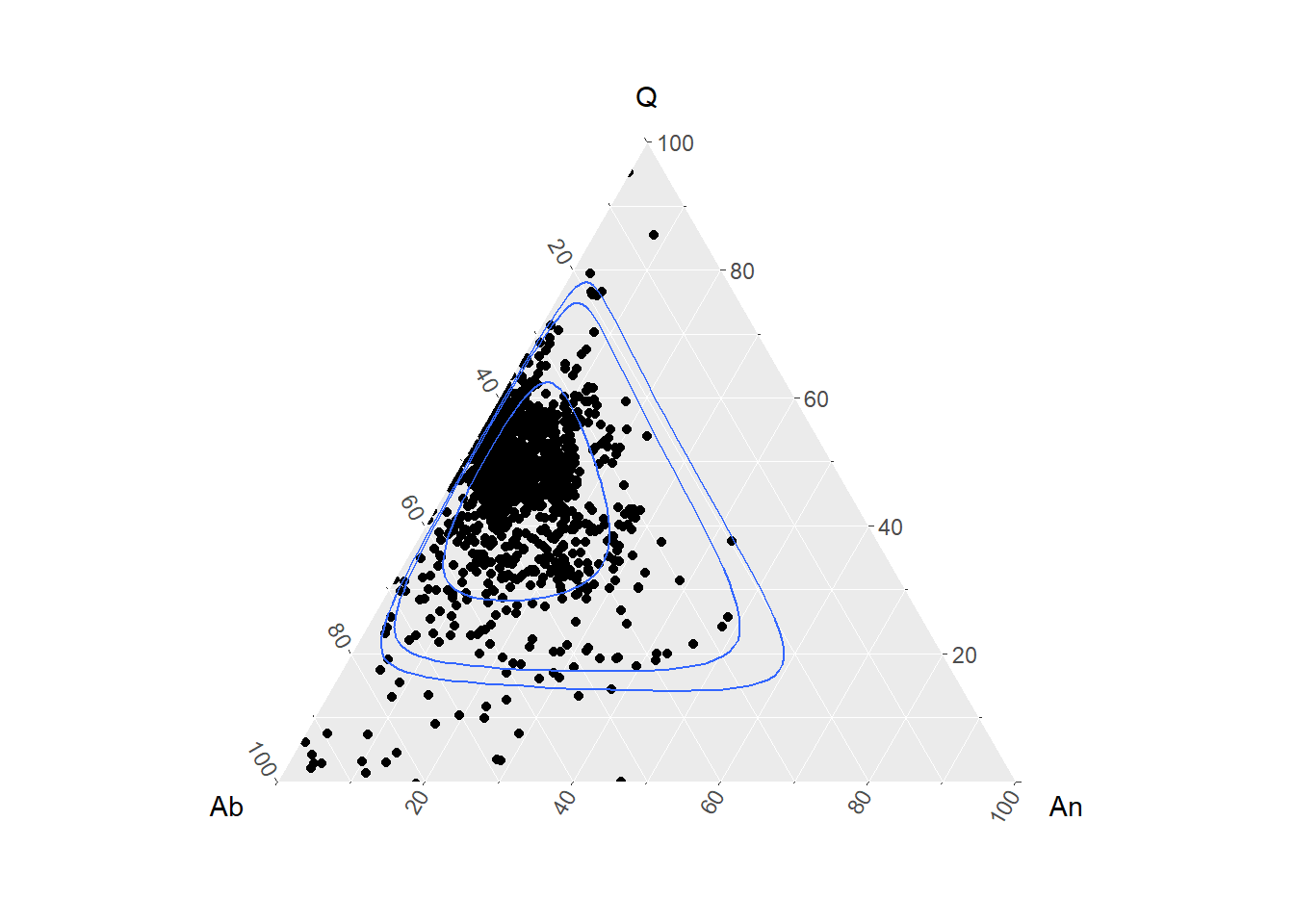
ou pivotado
BD %>%
filter(Tectonic_setting=="ARCHAEAN CRATONS") %>%
select(Q = Quartz,
Ab = Albite,
An = Anorthite,
SubGroup) %>%
ggtern(aes(x= Ab, y=Q, z=An, col = SubGroup)) +
geom_point() +
facet_wrap( ~ SubGroup)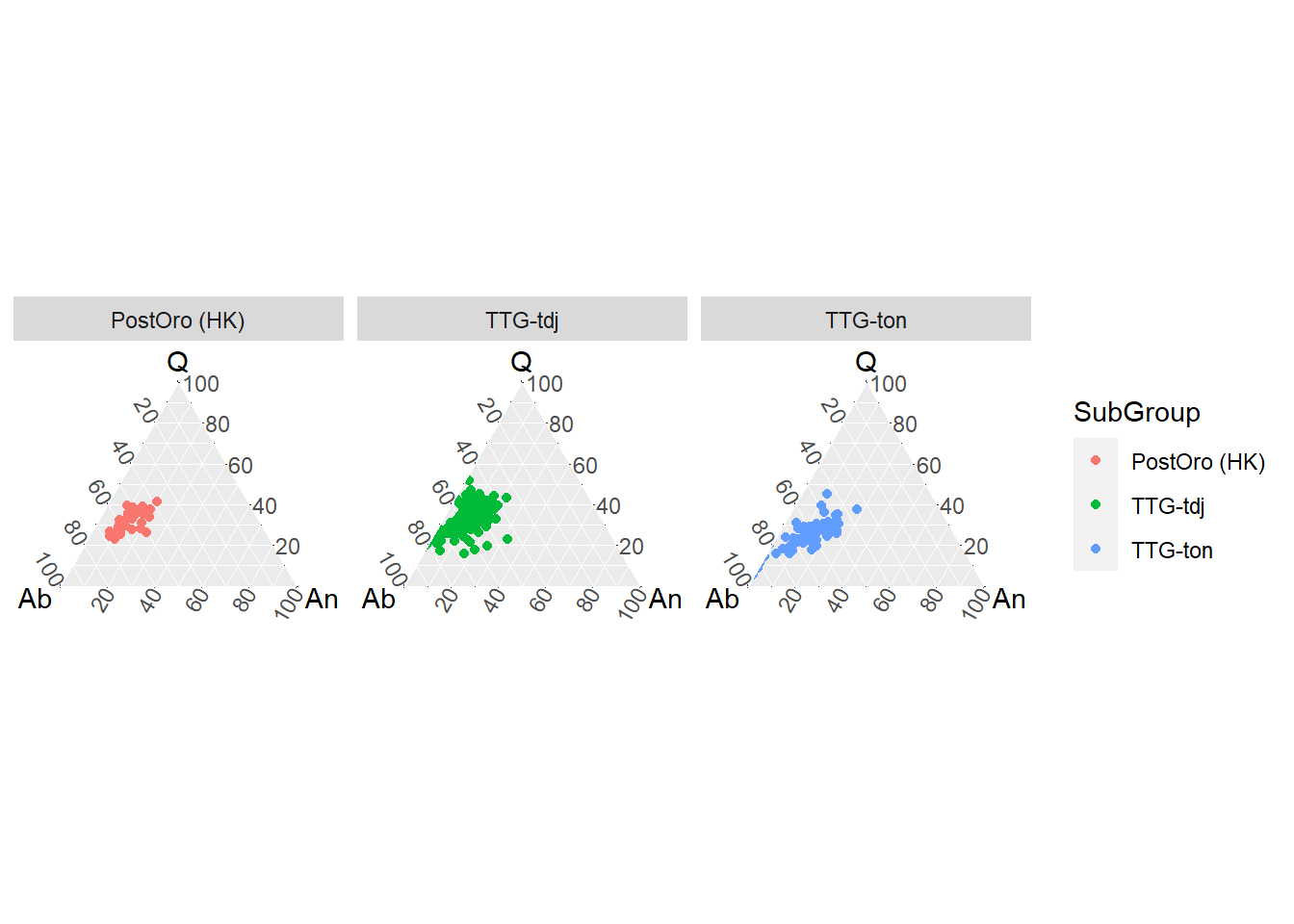
Barras
Horizontal
BD %>%
reframe(n=n(),
.by = Tectonic_setting) %>%
ggplot(aes(x=n,
y=fct_reorder(Tectonic_setting,n),
fill=Tectonic_setting))+
geom_bar(stat="identity")+
labs(y="Ambientes tectônicos") +
theme(axis.title.x = element_blank(),
legend.position = "none")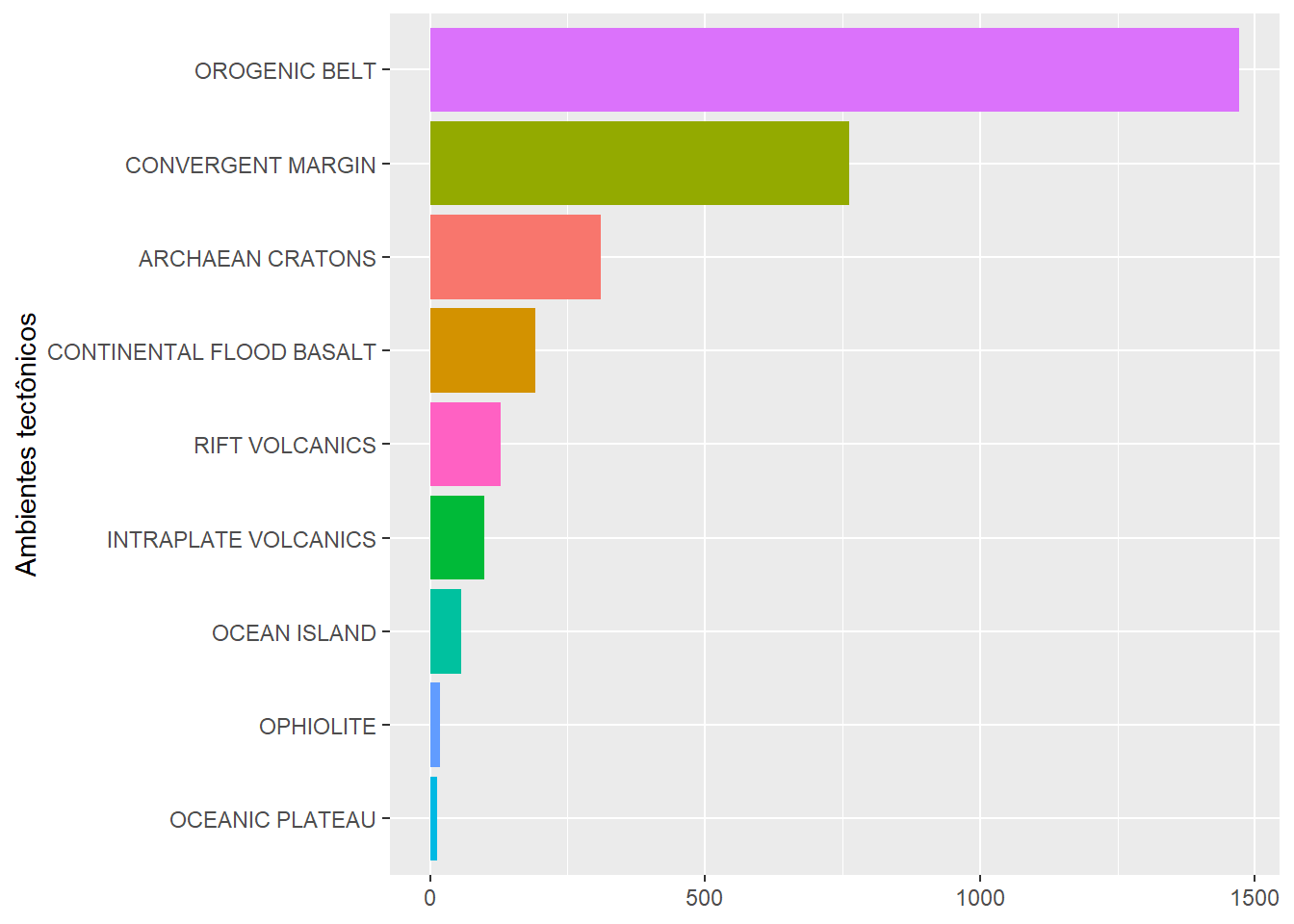
Vertical
BD %>%
reframe(n=n(),
.by = Tectonic_setting) %>%
ggplot(aes(x=fct_reorder(Tectonic_setting,desc(n)),
y=n,
fill=Tectonic_setting))+
geom_bar(stat="identity")+
scale_x_discrete(guide = guide_axis(n.dodge = 4))+
theme(axis.title.x = element_blank(),
legend.position = "none")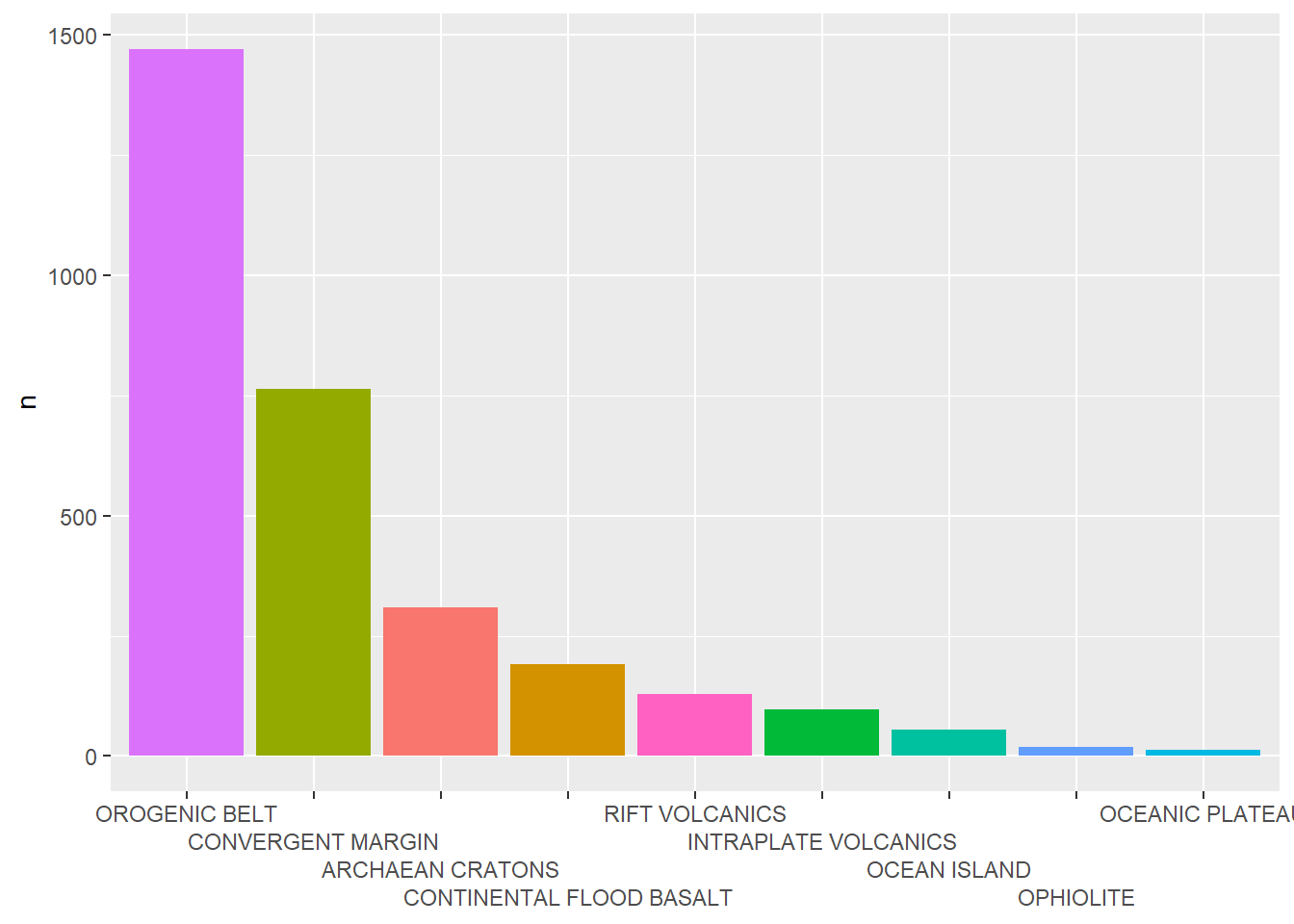
Pie-Chart
Simples, calculando a média com reframe() + mean()
BD %>%
select(
Q = Quartz,
Ab = Albite,
An = Anorthite,
Tectonic = Tectonic_setting
) %>%
reframe(
Q = mean(Q, na.rm = TRUE),
Ab = mean(Ab, na.rm = TRUE),
An = mean(An, na.rm = TRUE)
) %>%
pivot_longer(cols = Q:An,
names_to = "mineral",
values_to = "value") %>%
ggplot(aes(x = "", y = value, fill = mineral)) +
geom_bar(stat = "identity", width = 1) +
coord_polar("y")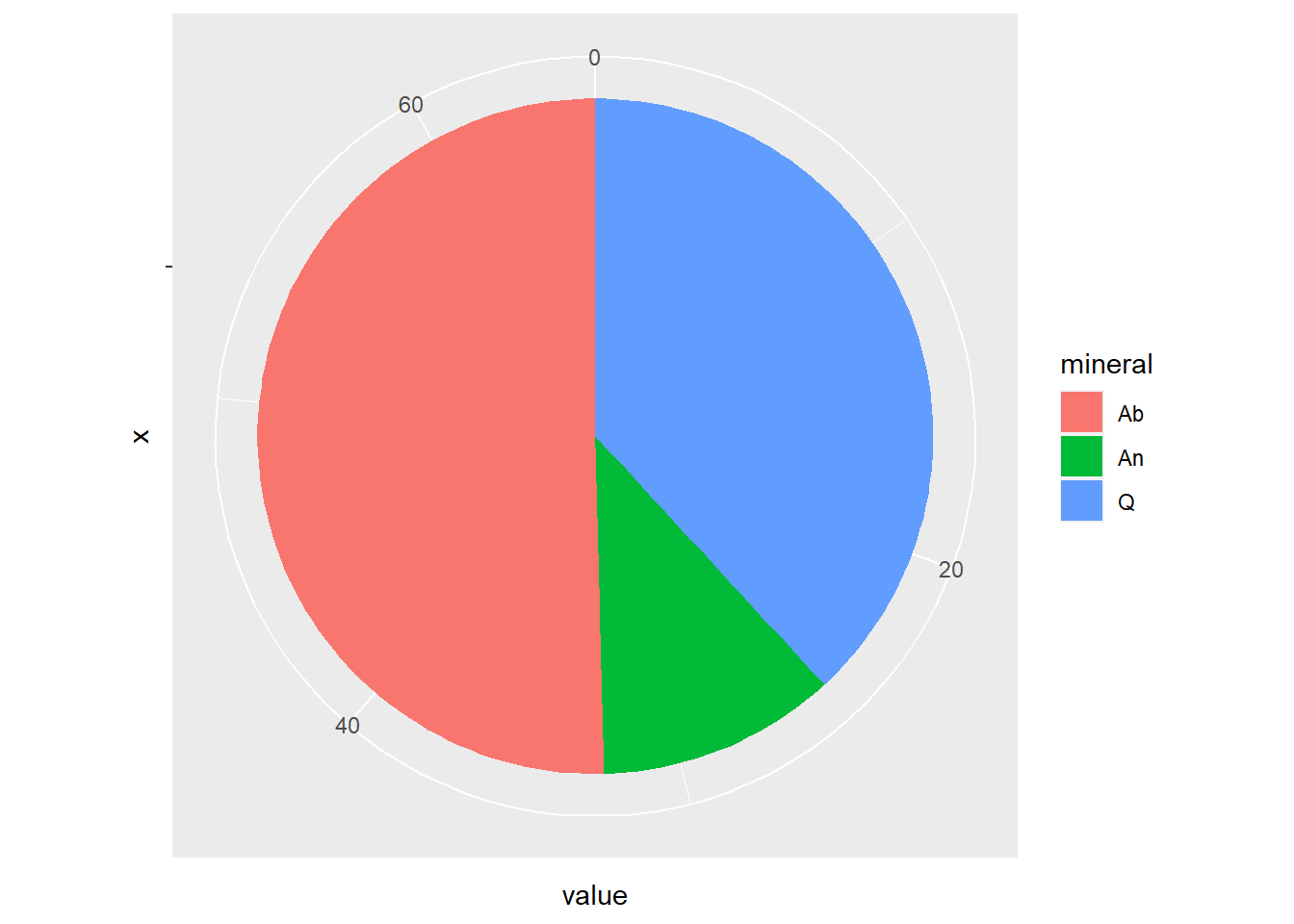
Ou então, pivotado
BD %>%
select(
Q = Quartz,
Ab = Albite,
Or = Orthoclase,
Tectonic = Tectonic_setting
) %>%
drop_na(Q,Ab,Or) %>%
reframe(
Q = mean(Q, na.rm = TRUE),
Ab = mean(Ab, na.rm = TRUE),
Or = mean(Or, na.rm = TRUE),
.by=Tectonic) %>%
group_by(Tectonic) %>%
mutate(total=sum(Q,Ab,Or),
across(.cols=Q:Or,~.x*100/total)) %>%
ungroup() %>%
pivot_longer(cols = Q:Or,
names_to = "mineral",
values_to = "value") %>%
ggplot(aes(x = "", y = value, fill = mineral)) +
geom_bar(stat = "identity", width = 1) +
coord_polar("y")+
facet_wrap(~Tectonic)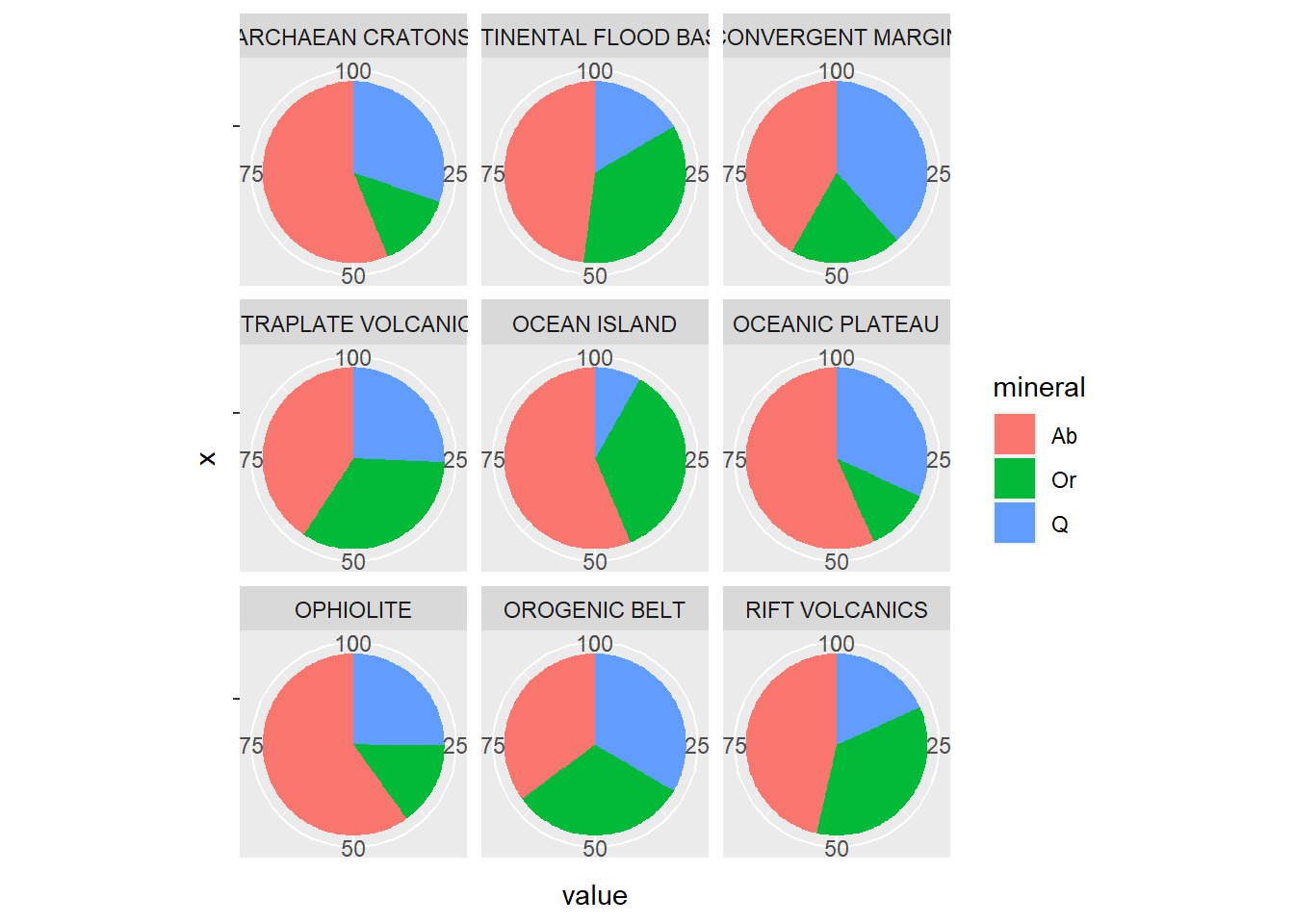
Direcional
Roseta simples
PALEOCURRENTS %>%
ggplot(aes(x=direction,fill=facies))+
geom_histogram(color="white",bins=30,breaks=seq(0, 360,by=10))+
coord_polar()+
scale_x_continuous("", limits = c(0, 360),
breaks = seq(0, 360,by=30),
labels = seq(0, 360,by=30))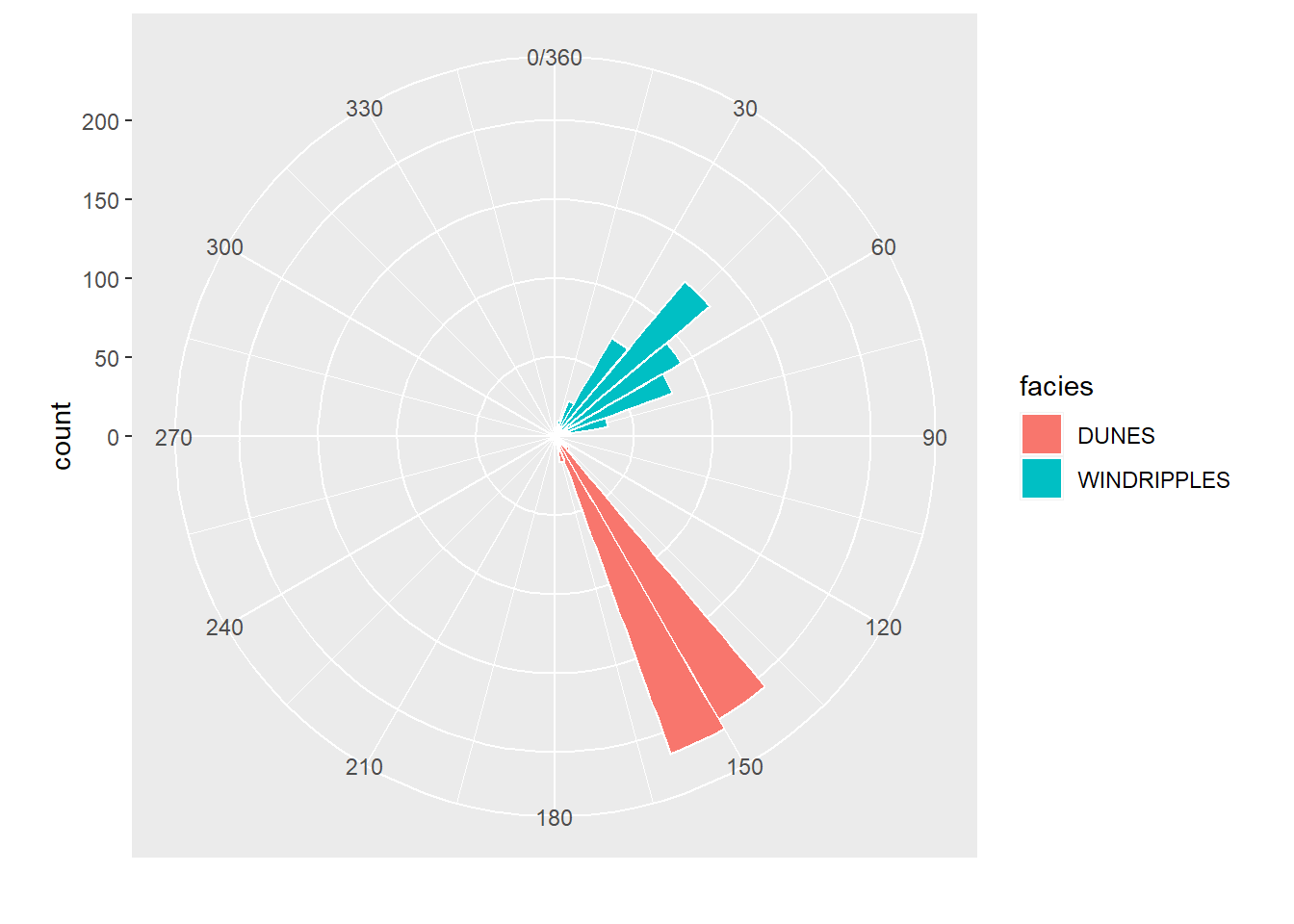
Ou facetado
PALEOCURRENTS %>%
ggplot(aes(x=direction,fill=facies))+
geom_histogram(color="white",bins=30,breaks=seq(0, 360,by=10))+
coord_polar()+
scale_x_continuous("", limits = c(0, 360),
breaks = seq(0, 360,by=30),
labels = seq(0, 360,by=30)) +
facet_wrap(~facies)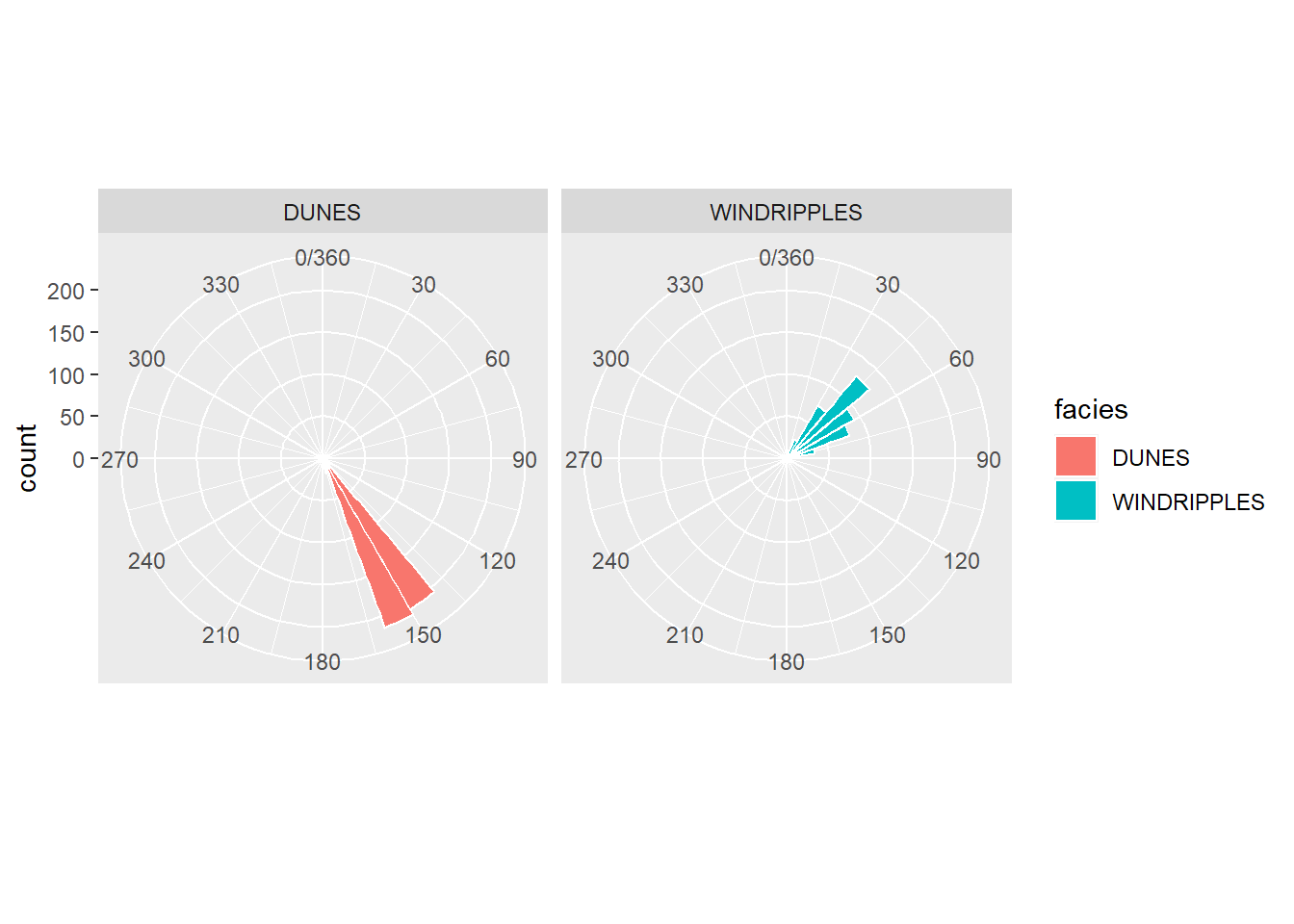
Estratigráfico
lvsd %>%
filter(well == "TG-227") %>%
pivot_longer(
cols = c(p_wave:s_wave, porosity, density),
names_to = "var",
values_to = "value"
) %>%
dplyr::mutate(var = factor(var, levels = c("porosity", "density", "p_wave", "s_wave"))) %>%
ggplot(aes(x = value, y = depth)) +
geom_rect(
mapping = aes(
ymin = 283.45,
ymax = 298.10,
xmin = -Inf,
xmax = Inf
),
alpha = 0.5,
fill = "gray",
inherit.aes = FALSE
) +
geom_rect(
mapping = aes(
ymin = 298.10,
ymax = 340,
xmin = -Inf,
xmax = Inf
),
alpha = 0.3,
fill = "lightyellow",
inherit.aes = FALSE
) +
geom_hline(
yintercept = c(283.45, 298.10, 340),
col = "red",
lty = 2,
alpha = 0.7
) +
geom_lineh() +
geom_point() +
scale_y_reverse() +
facet_geochem_gridh(vars(var), scales = "free") +
labs(x = NULL,
y = "Depth (m)") +
theme_paleo()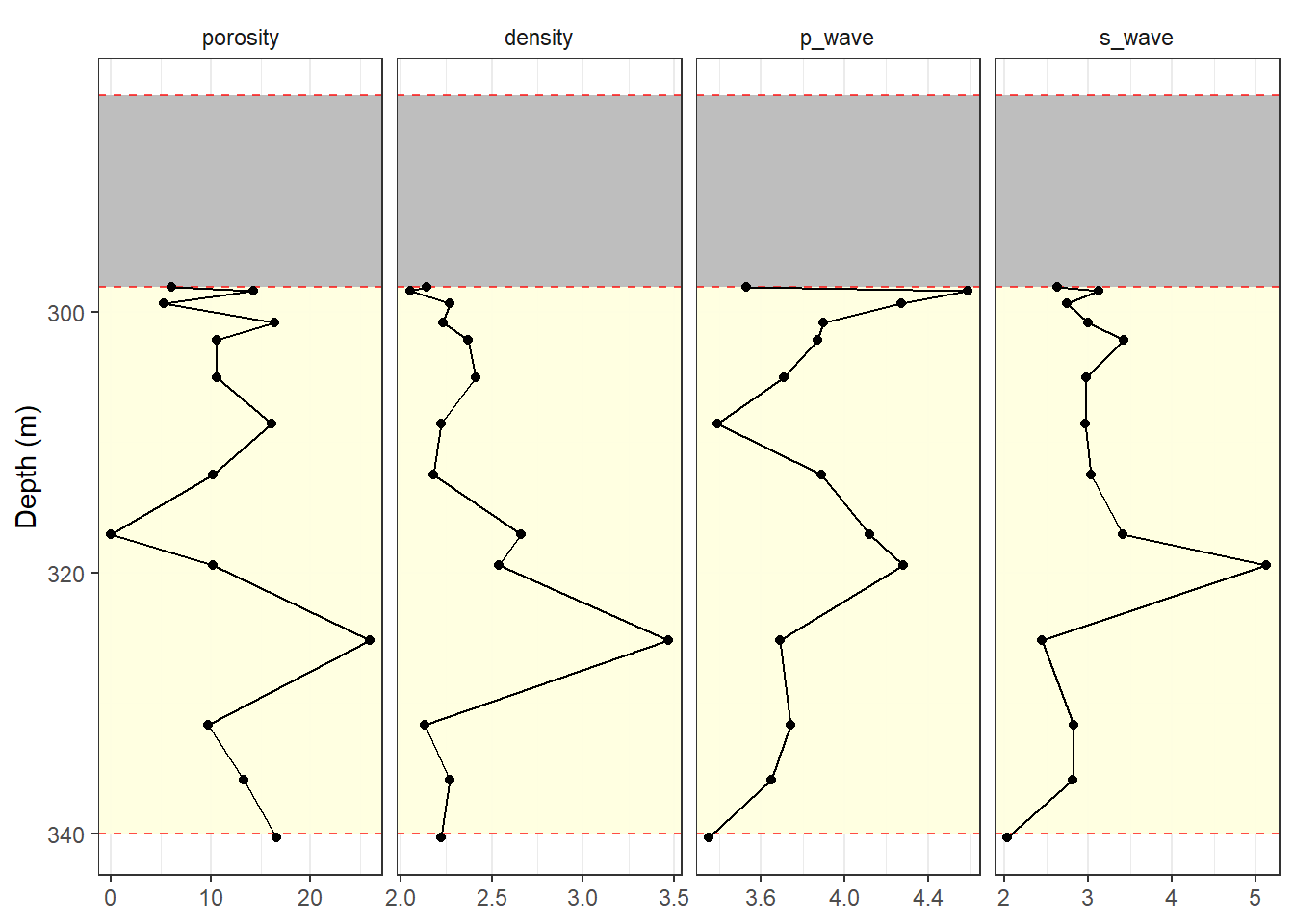
Mapas
Para usar o R para mapas, algumas bibliotecas são fundamentais, como:
- sf: Cria objetos espaciais com sistemas de referência de coordenadas e diversas funções de geoprocessamento.
- terra e raster: Pacotes indicados para trabalhar com dados do tipo raster, cada um com suas particularidades.
Outras bibliotecas são muito úteis para coisas em especifico
- chronosphere: Focado em dados de períodos geológicos distintos, com API para dados paleogeográficos e paleoclimáticos.
- mapview: Biblioteca para visualização rapida de dados espaciais de forma interativa
- ggspatial, ggdal, tidyterra: Extensões do
ggplot2para manipulação e visualização de dados espaciais. - geobr: Fornece bases cartográficas brasileiras.
- vapour: Leitura eficiente de arquivos raster, como imagens de satélite e DEMs.
A estrutura para mapas com ggplot é igual as anteriores. Vamos começar criando um banco de dados dummy com coordenadas dentro do brasil
coords_br <- data.frame(
id=1:10,
latitude = runif(10, min = -33, max = 0), # Valores aleatórios para latitude
longitude = runif(10, min = -54, max = -40) # Valores aleatórios para longitude
)
coords_br %>%
gt_preview()| id | latitude | longitude | |
|---|---|---|---|
| 1 | 1 | -23.639832 | -52.76330 |
| 2 | 2 | -32.875365 | -47.86329 |
| 3 | 3 | -15.526395 | -53.90008 |
| 4 | 4 | -19.012403 | -43.36837 |
| 5 | 5 | -19.344231 | -42.91496 |
| 6..9 | |||
| 10 | 10 | -3.356715 | -45.23681 |
Vamos visualizar rapidamente com mapview, mas para isso, primeiro precisamos converter o dataframe coords_br para um dado espacial. Para isso, vamos usar a função st_as_sf() da biblioteca sf:
# Coordinate Ref System WGS84 latlong = 4326
# Funciona melhor no viewer
library(sf)
coords_br %>%
st_as_sf(coords = c("longitude","latitude"), # informar nome colunas com X,Y
crs=4326) ->BDS_SPATIAL
BDS_SPATIAL %>% # informar crs WGS84 / EPSG = european petroleum society...
mapview(.)Vamos tilizar agora o pacote geobr para baixar dados cartograficos do brasil.
# baixar shapefile estados brasileiros
estados<-geobr::read_state()
POA<-geobr::read_municipality() %>%
filter(name_muni=="POA")
# filtrar para RS
rs <- estados %>%
filter(abbrev_state=="RS")
# criar coordenadas dummies dentro do RS
coords<-st_sample(rs, 100) Criação do mapa com ggplot: - Primeiro vamos fazer um mapa regional
inset<- ggplot()+
geom_sf(data = estados,fill="black")+
geom_sf(data = rs,fill="red")+
theme_void() + # Remove elementos do gráfico
theme(plot.background = element_rect(fill = "transparent", colour = NA)) # Define o fundo como transparente
inset
- Segundo seguimos com o mapa de interesse, no caso o estado do RS
RS<-ggplot()+
geom_sf(data = rs,linewidth=1)+
geom_sf(data = coords,color="grey50",size=2)+
geom_sf(data=POA,fill="red")+
coord_sf(xlim=c(-48,-59),
ylim=c(-26,-34))+
ggspatial::annotation_north_arrow(location="tr")+
ggspatial::annotation_scale(location="bl")
RS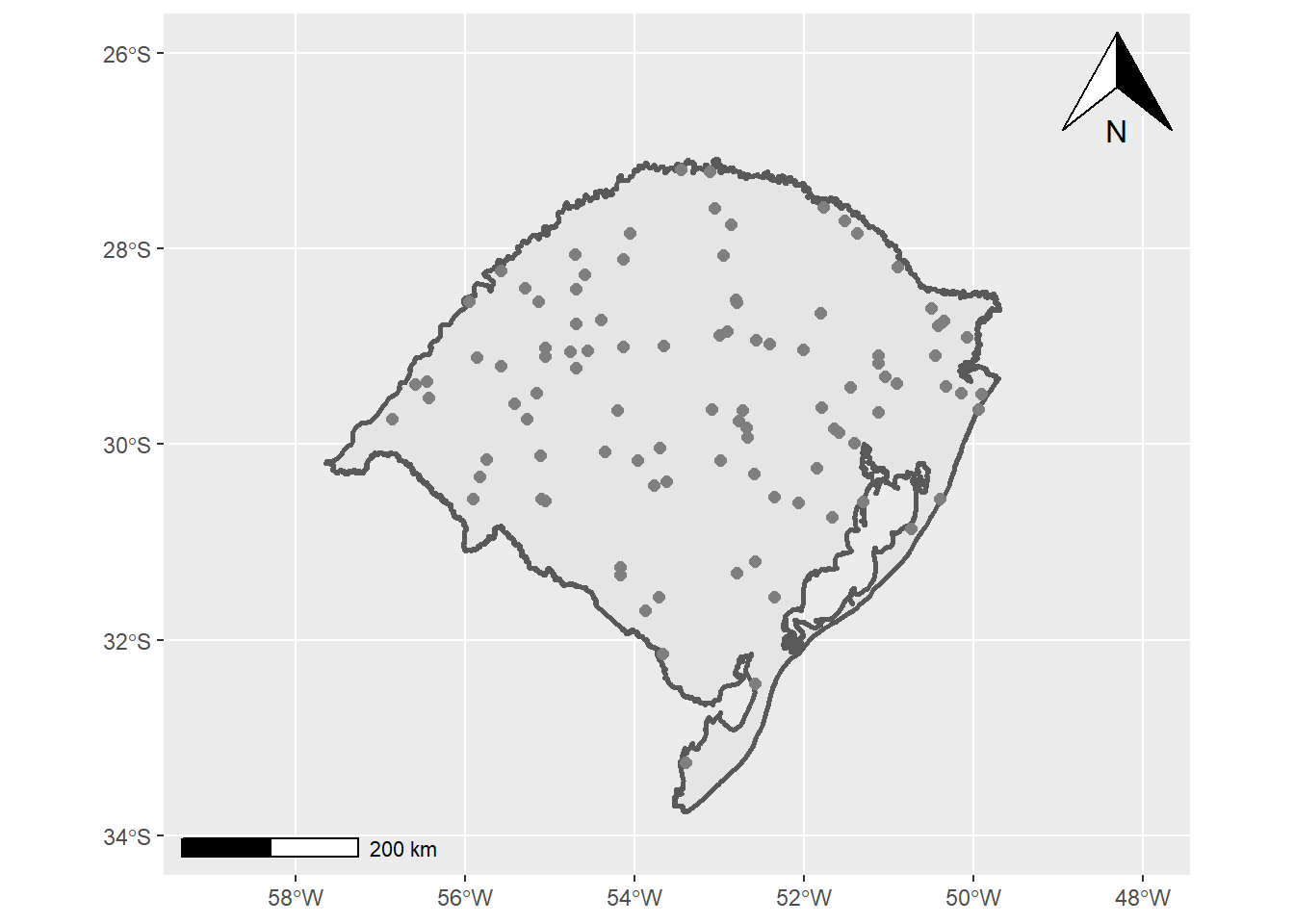
- 3 unir os plots com a biblioteca patchwork
#Patchwork
RS + inset_element(inset, left = 0, bottom = 0.7, right = 0.3, top = 1)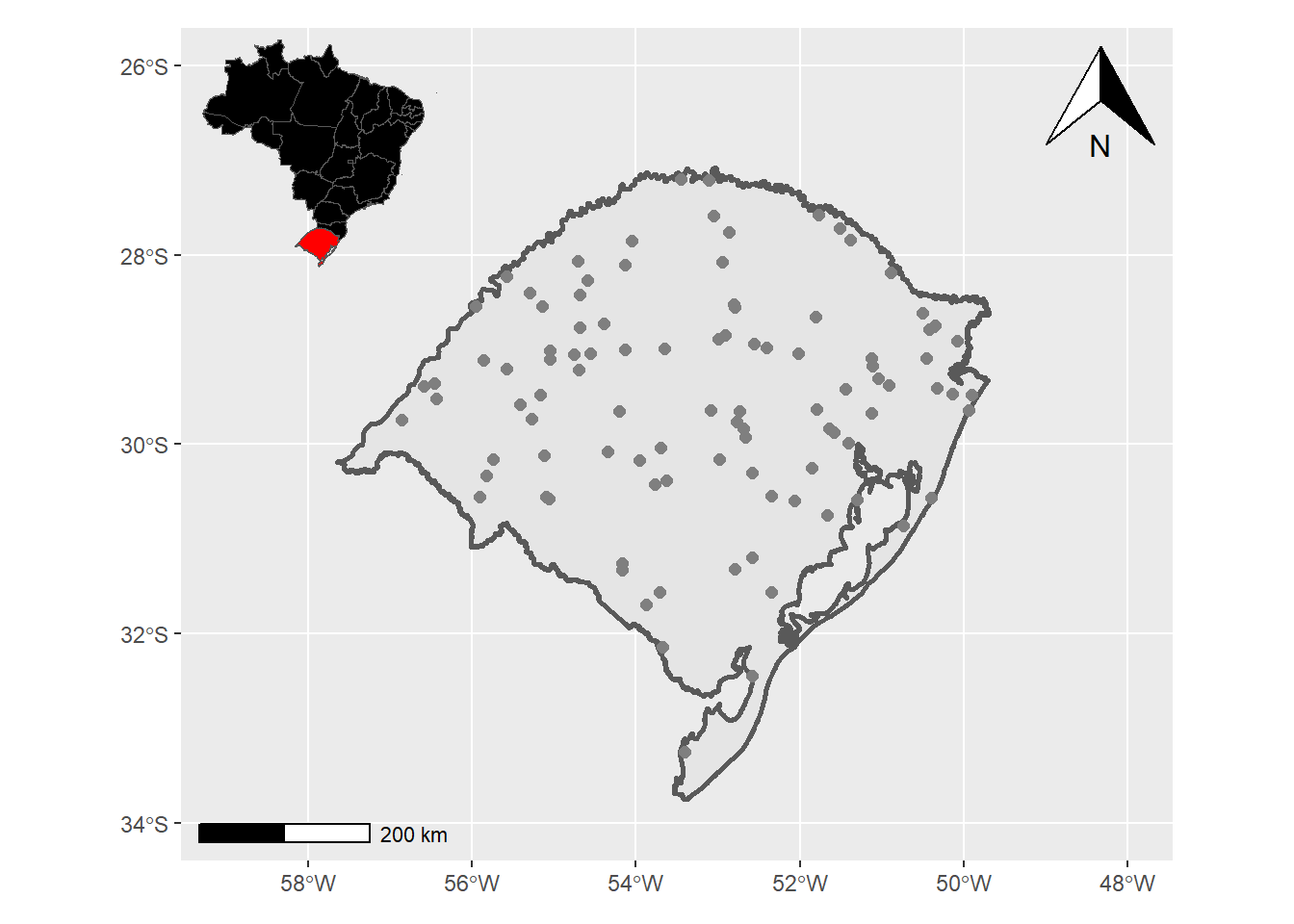
Outra opção, é utilziar o pacote ggdal para utilizar layers de imagem de satélites.
library(ggdal)
ggplot()+
ggdal::annotation_gdal(dsn = "virtualearth")+
geom_sf(data = rs,
linewidth=1.5,
fill=NA,col="white")+
geom_sf(data = coords,color="red",size=2)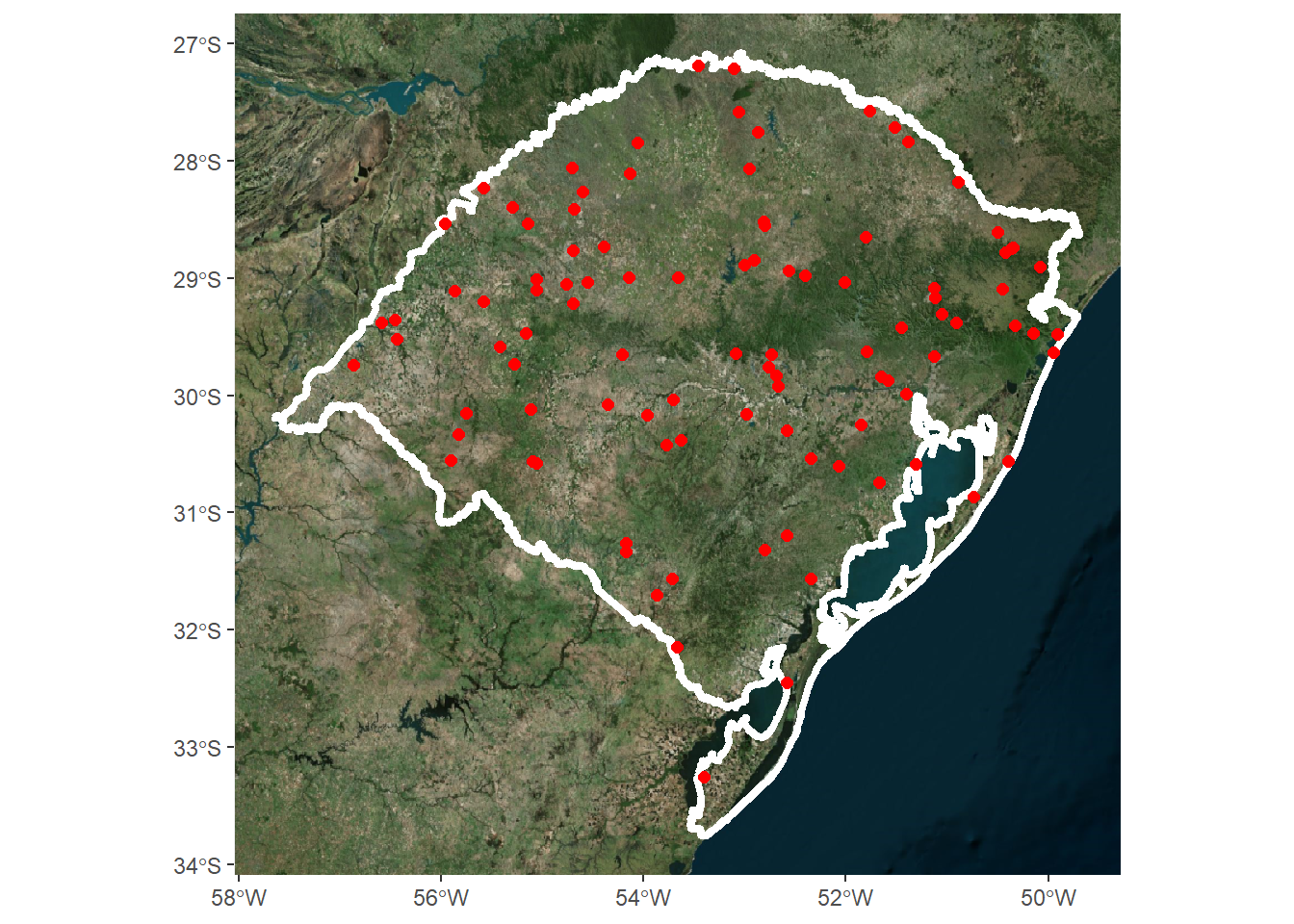
Cores
Cores podem inseridas através de:
Seus nomes (“white”, “aliceblue”, “antiquewhite”). Cheque todas usando a função colors()
colors()[1] "white" "aliceblue" "antiquewhite" [4] "antiquewhite1" "antiquewhite2" "antiquewhite3" [7] "antiquewhite4" "aquamarine" "aquamarine1" [10] "aquamarine2" "aquamarine3" "aquamarine4" [13] "azure" "azure1" "azure2" [16] "azure3" "azure4" "beige" [19] "bisque" "bisque1" "bisque2" [22] "bisque3" "bisque4" "black" [25] "blanchedalmond" "blue" "blue1" [28] "blue2" "blue3" "blue4" [31] "blueviolet" "brown" "brown1" [34] "brown2" "brown3" "brown4" [37] "burlywood" "burlywood1" "burlywood2" [40] "burlywood3" "burlywood4" "cadetblue" [43] "cadetblue1" "cadetblue2" "cadetblue3" [46] "cadetblue4" "chartreuse" "chartreuse1" [49] "chartreuse2" "chartreuse3" "chartreuse4" [52] "chocolate" "chocolate1" "chocolate2" [55] "chocolate3" "chocolate4" "coral" [58] "coral1" "coral2" "coral3" [61] "coral4" "cornflowerblue" "cornsilk" [64] "cornsilk1" "cornsilk2" "cornsilk3" [67] "cornsilk4" "cyan" "cyan1" [70] "cyan2" "cyan3" "cyan4" [73] "darkblue" "darkcyan" "darkgoldenrod" [76] "darkgoldenrod1" "darkgoldenrod2" "darkgoldenrod3" [79] "darkgoldenrod4" "darkgray" "darkgreen" [82] "darkgrey" "darkkhaki" "darkmagenta" [85] "darkolivegreen" "darkolivegreen1" "darkolivegreen2" [88] "darkolivegreen3" "darkolivegreen4" "darkorange" [91] "darkorange1" "darkorange2" "darkorange3" [94] "darkorange4" "darkorchid" "darkorchid1" [97] "darkorchid2" "darkorchid3" "darkorchid4" [100] "darkred" "darksalmon" "darkseagreen" [103] "darkseagreen1" "darkseagreen2" "darkseagreen3" [106] "darkseagreen4" "darkslateblue" "darkslategray" [109] "darkslategray1" "darkslategray2" "darkslategray3" [112] "darkslategray4" "darkslategrey" "darkturquoise" [115] "darkviolet" "deeppink" "deeppink1" [118] "deeppink2" "deeppink3" "deeppink4" [121] "deepskyblue" "deepskyblue1" "deepskyblue2" [124] "deepskyblue3" "deepskyblue4" "dimgray" [127] "dimgrey" "dodgerblue" "dodgerblue1" [130] "dodgerblue2" "dodgerblue3" "dodgerblue4" [133] "firebrick" "firebrick1" "firebrick2" [136] "firebrick3" "firebrick4" "floralwhite" [139] "forestgreen" "gainsboro" "ghostwhite" [142] "gold" "gold1" "gold2" [145] "gold3" "gold4" "goldenrod" [148] "goldenrod1" "goldenrod2" "goldenrod3" [151] "goldenrod4" "gray" "gray0" [154] "gray1" "gray2" "gray3" [157] "gray4" "gray5" "gray6" [160] "gray7" "gray8" "gray9" [163] "gray10" "gray11" "gray12" [166] "gray13" "gray14" "gray15" [169] "gray16" "gray17" "gray18" [172] "gray19" "gray20" "gray21" [175] "gray22" "gray23" "gray24" [178] "gray25" "gray26" "gray27" [181] "gray28" "gray29" "gray30" [184] "gray31" "gray32" "gray33" [187] "gray34" "gray35" "gray36" [190] "gray37" "gray38" "gray39" [193] "gray40" "gray41" "gray42" [196] "gray43" "gray44" "gray45" [199] "gray46" "gray47" "gray48" [202] "gray49" "gray50" "gray51" [205] "gray52" "gray53" "gray54" [208] "gray55" "gray56" "gray57" [211] "gray58" "gray59" "gray60" [214] "gray61" "gray62" "gray63" [217] "gray64" "gray65" "gray66" [220] "gray67" "gray68" "gray69" [223] "gray70" "gray71" "gray72" [226] "gray73" "gray74" "gray75" [229] "gray76" "gray77" "gray78" [232] "gray79" "gray80" "gray81" [235] "gray82" "gray83" "gray84" [238] "gray85" "gray86" "gray87" [241] "gray88" "gray89" "gray90" [244] "gray91" "gray92" "gray93" [247] "gray94" "gray95" "gray96" [250] "gray97" "gray98" "gray99" [253] "gray100" "green" "green1" [256] "green2" "green3" "green4" [259] "greenyellow" "grey" "grey0" [262] "grey1" "grey2" "grey3" [265] "grey4" "grey5" "grey6" [268] "grey7" "grey8" "grey9" [271] "grey10" "grey11" "grey12" [274] "grey13" "grey14" "grey15" [277] "grey16" "grey17" "grey18" [280] "grey19" "grey20" "grey21" [283] "grey22" "grey23" "grey24" [286] "grey25" "grey26" "grey27" [289] "grey28" "grey29" "grey30" [292] "grey31" "grey32" "grey33" [295] "grey34" "grey35" "grey36" [298] "grey37" "grey38" "grey39" [301] "grey40" "grey41" "grey42" [304] "grey43" "grey44" "grey45" [307] "grey46" "grey47" "grey48" [310] "grey49" "grey50" "grey51" [313] "grey52" "grey53" "grey54" [316] "grey55" "grey56" "grey57" [319] "grey58" "grey59" "grey60" [322] "grey61" "grey62" "grey63" [325] "grey64" "grey65" "grey66" [328] "grey67" "grey68" "grey69" [331] "grey70" "grey71" "grey72" [334] "grey73" "grey74" "grey75" [337] "grey76" "grey77" "grey78" [340] "grey79" "grey80" "grey81" [343] "grey82" "grey83" "grey84" [346] "grey85" "grey86" "grey87" [349] "grey88" "grey89" "grey90" [352] "grey91" "grey92" "grey93" [355] "grey94" "grey95" "grey96" [358] "grey97" "grey98" "grey99" [361] "grey100" "honeydew" "honeydew1" [364] "honeydew2" "honeydew3" "honeydew4" [367] "hotpink" "hotpink1" "hotpink2" [370] "hotpink3" "hotpink4" "indianred" [373] "indianred1" "indianred2" "indianred3" [376] "indianred4" "ivory" "ivory1" [379] "ivory2" "ivory3" "ivory4" [382] "khaki" "khaki1" "khaki2" [385] "khaki3" "khaki4" "lavender" [388] "lavenderblush" "lavenderblush1" "lavenderblush2" [391] "lavenderblush3" "lavenderblush4" "lawngreen" [394] "lemonchiffon" "lemonchiffon1" "lemonchiffon2" [397] "lemonchiffon3" "lemonchiffon4" "lightblue" [400] "lightblue1" "lightblue2" "lightblue3" [403] "lightblue4" "lightcoral" "lightcyan" [406] "lightcyan1" "lightcyan2" "lightcyan3" [409] "lightcyan4" "lightgoldenrod" "lightgoldenrod1" [412] "lightgoldenrod2" "lightgoldenrod3" "lightgoldenrod4" [415] "lightgoldenrodyellow" "lightgray" "lightgreen" [418] "lightgrey" "lightpink" "lightpink1" [421] "lightpink2" "lightpink3" "lightpink4" [424] "lightsalmon" "lightsalmon1" "lightsalmon2" [427] "lightsalmon3" "lightsalmon4" "lightseagreen" [430] "lightskyblue" "lightskyblue1" "lightskyblue2" [433] "lightskyblue3" "lightskyblue4" "lightslateblue" [436] "lightslategray" "lightslategrey" "lightsteelblue" [439] "lightsteelblue1" "lightsteelblue2" "lightsteelblue3" [442] "lightsteelblue4" "lightyellow" "lightyellow1" [445] "lightyellow2" "lightyellow3" "lightyellow4" [448] "limegreen" "linen" "magenta" [451] "magenta1" "magenta2" "magenta3" [454] "magenta4" "maroon" "maroon1" [457] "maroon2" "maroon3" "maroon4" [460] "mediumaquamarine" "mediumblue" "mediumorchid" [463] "mediumorchid1" "mediumorchid2" "mediumorchid3" [466] "mediumorchid4" "mediumpurple" "mediumpurple1" [469] "mediumpurple2" "mediumpurple3" "mediumpurple4" [472] "mediumseagreen" "mediumslateblue" "mediumspringgreen" [475] "mediumturquoise" "mediumvioletred" "midnightblue" [478] "mintcream" "mistyrose" "mistyrose1" [481] "mistyrose2" "mistyrose3" "mistyrose4" [484] "moccasin" "navajowhite" "navajowhite1" [487] "navajowhite2" "navajowhite3" "navajowhite4" [490] "navy" "navyblue" "oldlace" [493] "olivedrab" "olivedrab1" "olivedrab2" [496] "olivedrab3" "olivedrab4" "orange" [499] "orange1" "orange2" "orange3" [502] "orange4" "orangered" "orangered1" [505] "orangered2" "orangered3" "orangered4" [508] "orchid" "orchid1" "orchid2" [511] "orchid3" "orchid4" "palegoldenrod" [514] "palegreen" "palegreen1" "palegreen2" [517] "palegreen3" "palegreen4" "paleturquoise" [520] "paleturquoise1" "paleturquoise2" "paleturquoise3" [523] "paleturquoise4" "palevioletred" "palevioletred1" [526] "palevioletred2" "palevioletred3" "palevioletred4" [529] "papayawhip" "peachpuff" "peachpuff1" [532] "peachpuff2" "peachpuff3" "peachpuff4" [535] "peru" "pink" "pink1" [538] "pink2" "pink3" "pink4" [541] "plum" "plum1" "plum2" [544] "plum3" "plum4" "powderblue" [547] "purple" "purple1" "purple2" [550] "purple3" "purple4" "red" [553] "red1" "red2" "red3" [556] "red4" "rosybrown" "rosybrown1" [559] "rosybrown2" "rosybrown3" "rosybrown4" [562] "royalblue" "royalblue1" "royalblue2" [565] "royalblue3" "royalblue4" "saddlebrown" [568] "salmon" "salmon1" "salmon2" [571] "salmon3" "salmon4" "sandybrown" [574] "seagreen" "seagreen1" "seagreen2" [577] "seagreen3" "seagreen4" "seashell" [580] "seashell1" "seashell2" "seashell3" [583] "seashell4" "sienna" "sienna1" [586] "sienna2" "sienna3" "sienna4" [589] "skyblue" "skyblue1" "skyblue2" [592] "skyblue3" "skyblue4" "slateblue" [595] "slateblue1" "slateblue2" "slateblue3" [598] "slateblue4" "slategray" "slategray1" [601] "slategray2" "slategray3" "slategray4" [604] "slategrey" "snow" "snow1" [607] "snow2" "snow3" "snow4" [610] "springgreen" "springgreen1" "springgreen2" [613] "springgreen3" "springgreen4" "steelblue" [616] "steelblue1" "steelblue2" "steelblue3" [619] "steelblue4" "tan" "tan1" [622] "tan2" "tan3" "tan4" [625] "thistle" "thistle1" "thistle2" [628] "thistle3" "thistle4" "tomato" [631] "tomato1" "tomato2" "tomato3" [634] "tomato4" "turquoise" "turquoise1" [637] "turquoise2" "turquoise3" "turquoise4" [640] "violet" "violetred" "violetred1" [643] "violetred2" "violetred3" "violetred4" [646] "wheat" "wheat1" "wheat2" [649] "wheat3" "wheat4" "whitesmoke" [652] "yellow" "yellow1" "yellow2" [655] "yellow3" "yellow4" "yellowgreen"Por seus códigos
par(mfrow=c(1,2)) # Hexadecimal plot(1:10, col = "#FF5733", main="Hexadecimal") # RGB plot(1:10, col = rgb(255, 87, 51, maxColorValue = 255),main="RGB")
par(mfrow=c(1,1))Paletas prontas de cores- as mais utilizadas são Viridis e rColorBrewer
As paletas de cores são em geral convergentes, divergentes ou categóricas. Outra subdivisão são valores continuos ou discretos.
Viridis
Scale_*_viridis_b
Usar scale_*_viridis_b para dados continuos separados em grupos (bins). * representa ou color ou fill.
BD %>%
ggplot(aes(x=SiO2,
y=TiO2,
color=Al2O3))+
geom_point()+
scale_color_viridis_b(option = "magma",n.breaks=3) +
labs(title = "Viridis - Magma (A)")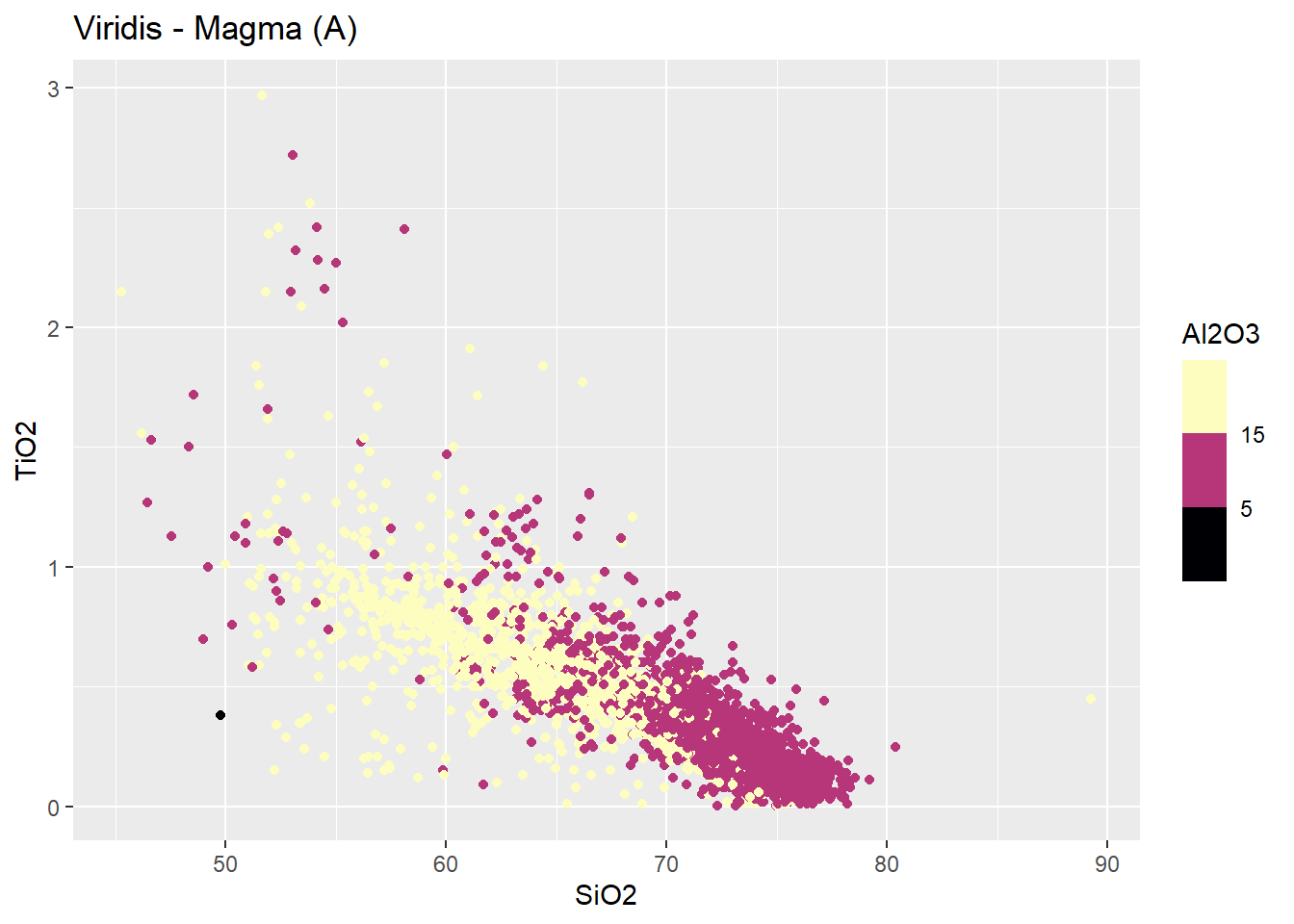
Scale_*_viridis_c
Usar scale_*_viridis_c para dados continuos. * representa ou color ou fill : color para as linhas (no caso o ponto) e fill para colunas.
BD %>%
ggplot(aes(x=SiO2,
y=TiO2,
color=Al2O3))+
geom_point()+
scale_color_viridis_c(option = "magma") +
labs(title = "Viridis - Magma (A)")->A
BD %>%
ggplot(aes(x=SiO2,
y=TiO2,
color=Al2O3))+
geom_point()+
scale_color_viridis_c(option = "inferno") +
labs(title = "Viridis - Inferno (B)")->B
BD %>%
ggplot(aes(x=SiO2,
y=TiO2,
color=Al2O3))+
geom_point()+
scale_color_viridis_c(option = "plasma") +
labs(title = "Viridis - Plasma (C)")->C
BD %>%
ggplot(aes(x=SiO2,
y=TiO2,
color=Al2O3))+
geom_point()+
scale_color_viridis_c(option = "viridis") +
labs(title = "Viridis - Viridis (D)")->D
BD %>%
ggplot(aes(x=SiO2,
y=TiO2,
color=Al2O3))+
geom_point()+
scale_color_viridis_c(option = "cividis") +
labs(title = "Viridis - Cividis (E)")->E
BD %>%
ggplot(aes(x=SiO2,
y=TiO2,
color=Al2O3))+
geom_point()+
scale_color_viridis_c(option = "rocket") +
labs(title = "Viridis - rocket (F)")->F1
BD %>%
ggplot(aes(x=SiO2,
y=TiO2,
color=Al2O3))+
geom_point()+
scale_color_viridis_c(option = "mako") +
labs(title = "Viridis - Mako (G)")->G
BD %>%
ggplot(aes(x=SiO2,
y=TiO2,
color=Al2O3))+
geom_point()+
scale_color_viridis_c(option = "turbo") +
labs(title = "Viridis - Turbo(H)")->H
A+B+C+D+E+F1+G+H+
plot_layout(ncol = 2)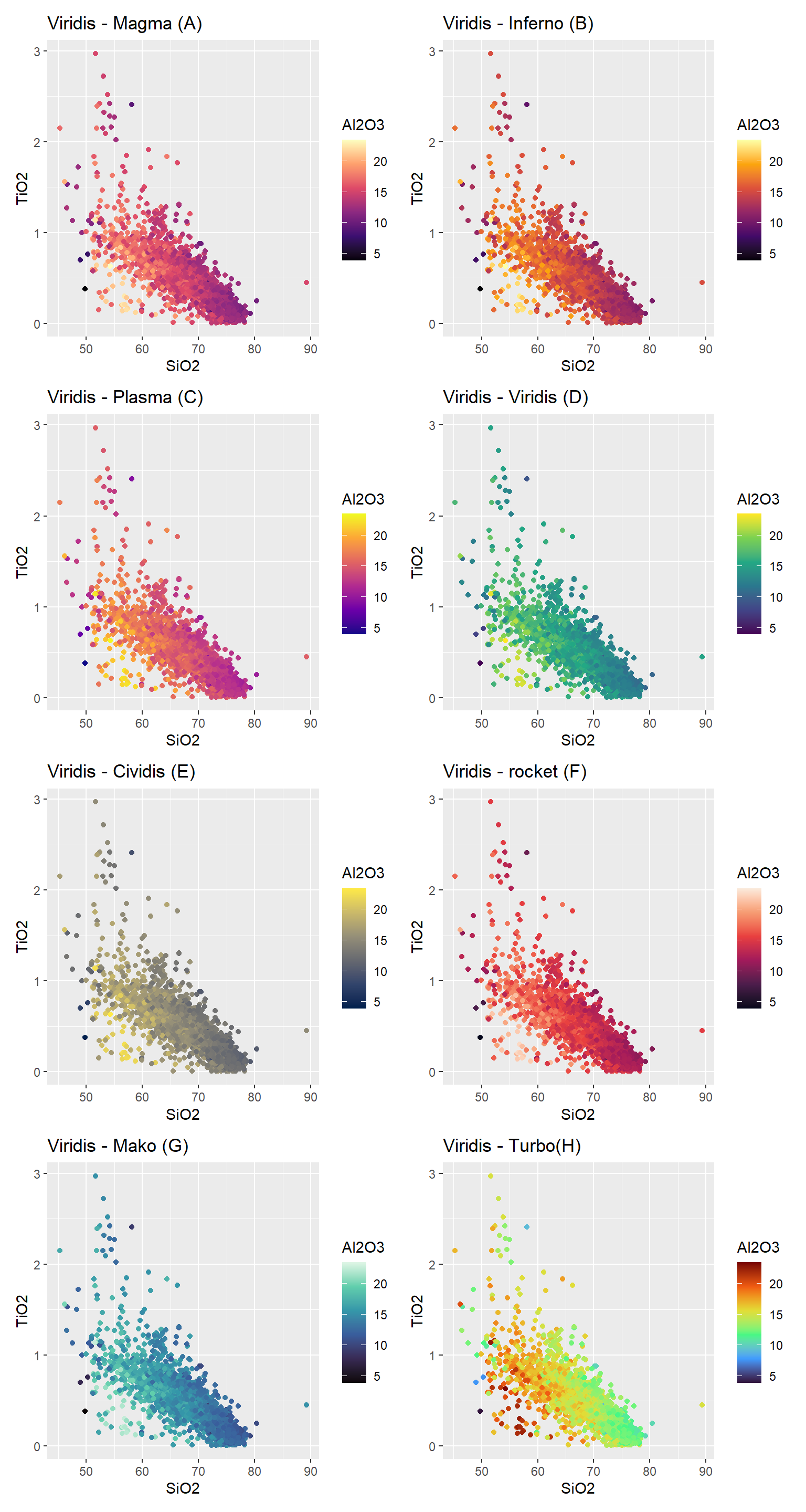
Scale_*_viridis_d
Usar scale_*_viridis_dpara dados discretos. * representa ou color ou fill : color para as linhas (no caso o ponto) e fill para colunas.
BD %>%
ggplot(aes(x=fct_reorder(Tectonic_setting,TiO2),
y=TiO2,
fill=fct_reorder(Tectonic_setting,TiO2)))+
geom_boxplot()+
scale_fill_viridis_d(option = "magma") +
labs(title = "Viridis - Magma (A)",
fill="Ambiente Tectônico")+
theme(axis.title.x=element_blank(), # retirar nome escala
axis.text.x=element_blank(), # retirar texto escala
axis.ticks.x=element_blank()) # retirar ticks escala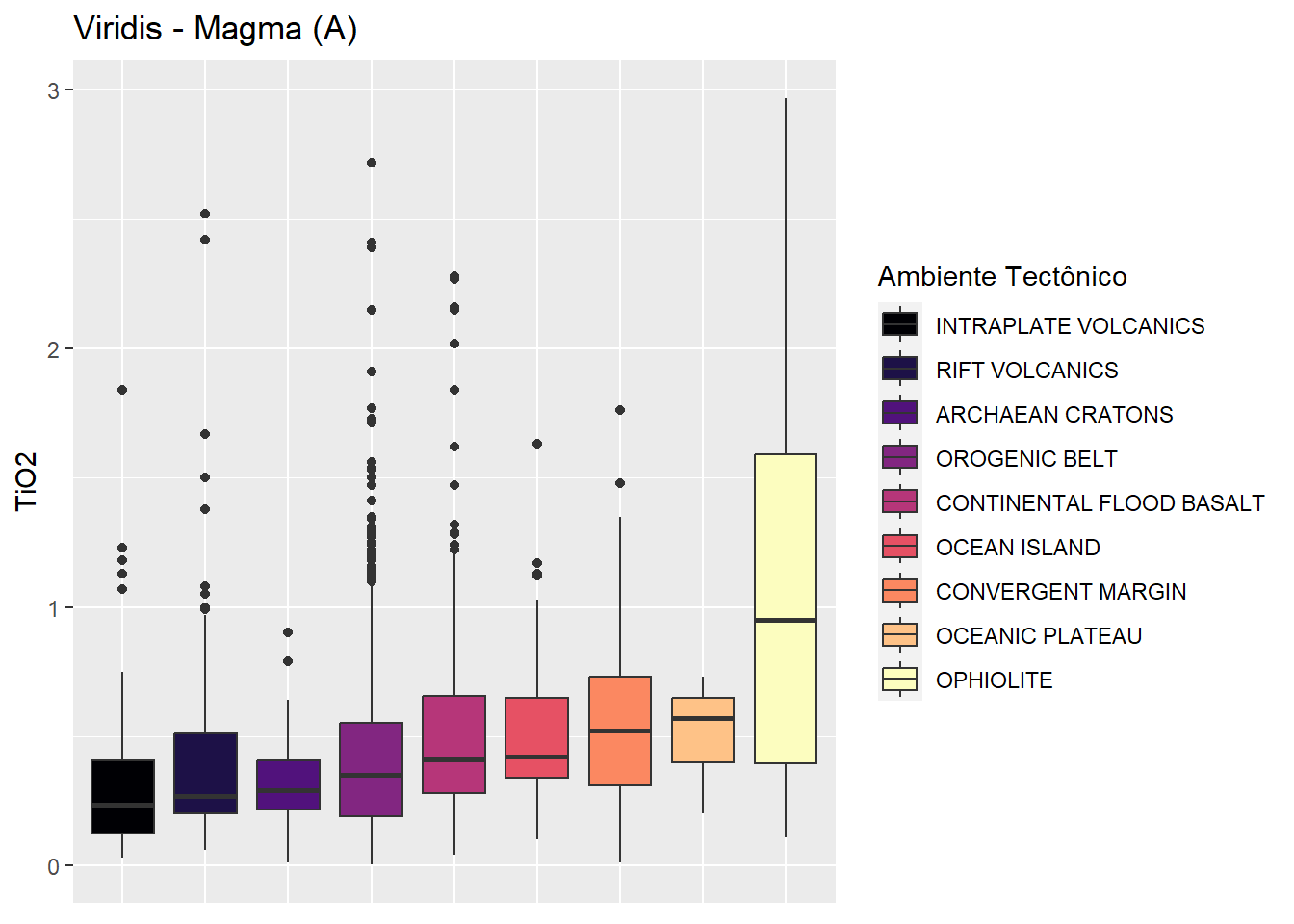
ColorBrew
Paleta de cores “color-blind friendly” facilitando visualização universal de gráficos. O conjunto completo de cores pode ser obtido com a função display.brewer.all().
display.brewer.all()
A biblioteca colorbrewer divide suas pallete (conjunto de cores) em sequencial, divergente e categórico, usando a função scale_*_brewer() e o parâmetro pallete para determinar pallete.
BD %>%
drop_na(CaO,Na2O,MgO) %>%
ggplot(aes(x=CaO,
y=Na2O,
colour=MgO))+
geom_point()+
scale_color_distiller(palette = "Greens",direction = 1) +
labs(title = "Sequencial Greens") -> A
BD %>%
drop_na(CaO,Na2O,MgO) %>%
ggplot(aes(x=CaO,
y=Na2O,
colour=MgO))+
geom_point()+
scale_color_distiller(palette = "YlOrRd",direction = 1) +
labs(title = "Sequencial YlOrRd") -> B
BD %>%
drop_na(Cu) %>%
ggplot(aes(x=fct_reorder(Tectonic_setting,Cu),
y=Cu,
fill=fct_reorder(Tectonic_setting,Cu)))+
geom_boxplot()+
scale_fill_brewer(palette = "Set1",direction = 1) +
scale_y_log10()+
labs(title = "Categórico Set1",
fill="Ambiente\nTectônico")+
theme(axis.title.x=element_blank(), # retirar nome escala
axis.text.x=element_blank(), # retirar texto escala
axis.ticks.x=element_blank())->C # retirar ticks escala
BD %>%
drop_na(Cu) %>%
ggplot(aes(x=fct_reorder(Tectonic_setting,Cu),
y=Cu,
fill=fct_reorder(Tectonic_setting,Cu)))+
geom_violin()+
scale_fill_brewer(palette = "Pastel1",direction = 1) +
scale_y_log10()+
labs(title = "Categórico Pastel1",
fill="Ambiente\nTectônico")+
theme(axis.title.x=element_blank(), # retirar nome escala
axis.text.x=element_blank(), # retirar texto escala
axis.ticks.x=element_blank())->D # retirar ticks escala
BD %>%
drop_na(Cu) %>%
ggplot(aes(x=SiO2,y=Al2O3,colour=log10(Cu)))+
geom_point()+
scale_colour_distiller(palette = "Spectral") +
labs(title = "Divergente Spectral",
fill="log10\nCobre")->E
BD %>%
drop_na(FeOt) %>%
filter(FeOt>0) %>%
ggplot(aes(x=SiO2,y=Al2O3,colour=FeOt))+
geom_point()+
scale_colour_distiller(palette = "RdBu") +
labs(title = "Divergente RdBu",
fill="Ferro\ntotal")->F1
A+B+C+D+E+F1+plot_layout(ncol=1)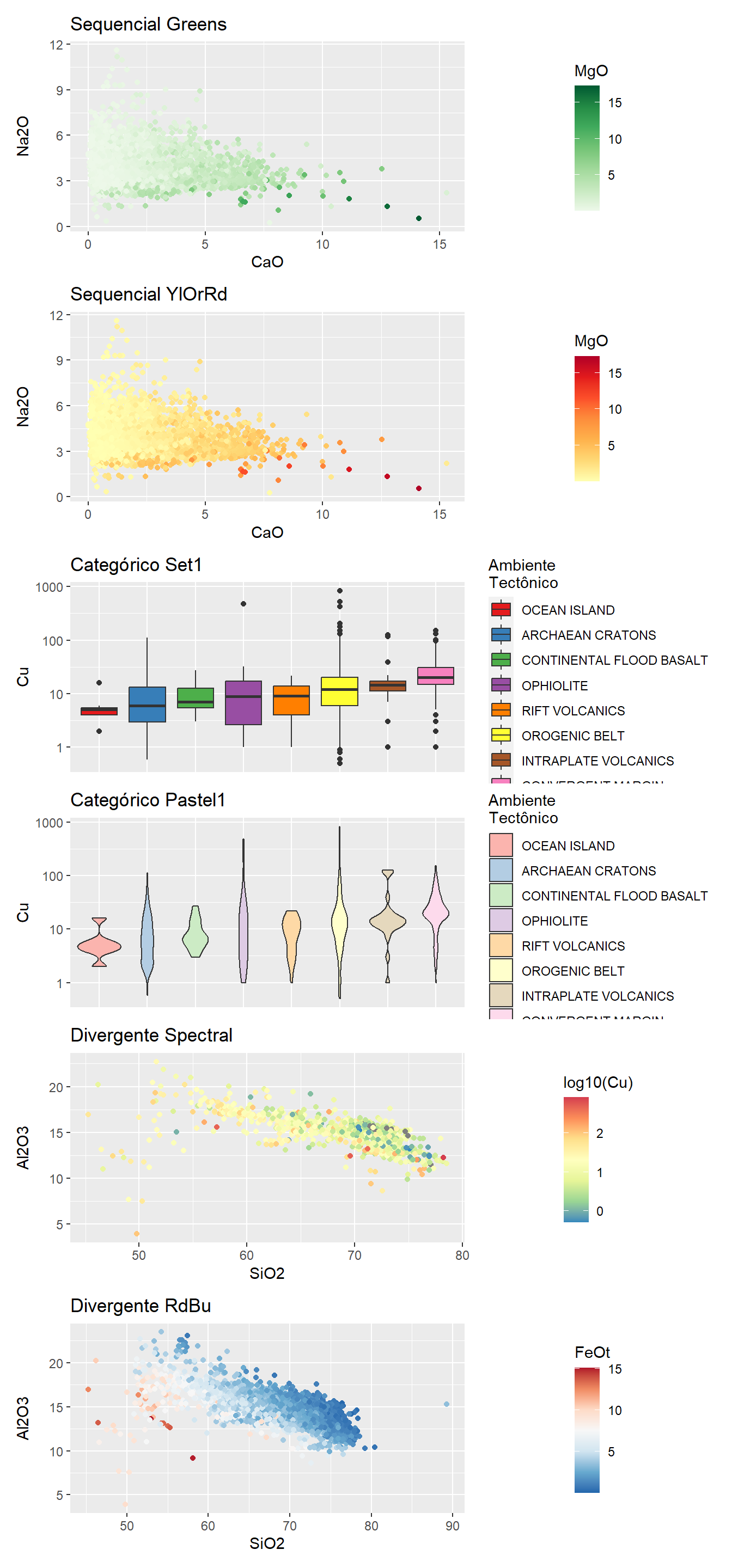
Cores manuais
No banco de dados
BD %>%
select(Tectonic_setting,TiO2,Na2O) %>%
#
mutate(colors=case_when(
Tectonic_setting=="CONVERGENT MARGIN"~"red",
Tectonic_setting=="ARCHAEAN CRATONS"~"blue",
Tectonic_setting=="OROGENIC BELT"~"yellow",
TRUE~"black"
)) %>%
ggplot(aes(x=TiO2,y=Na2O,color=colors))+
geom_point()+
scale_color_identity() 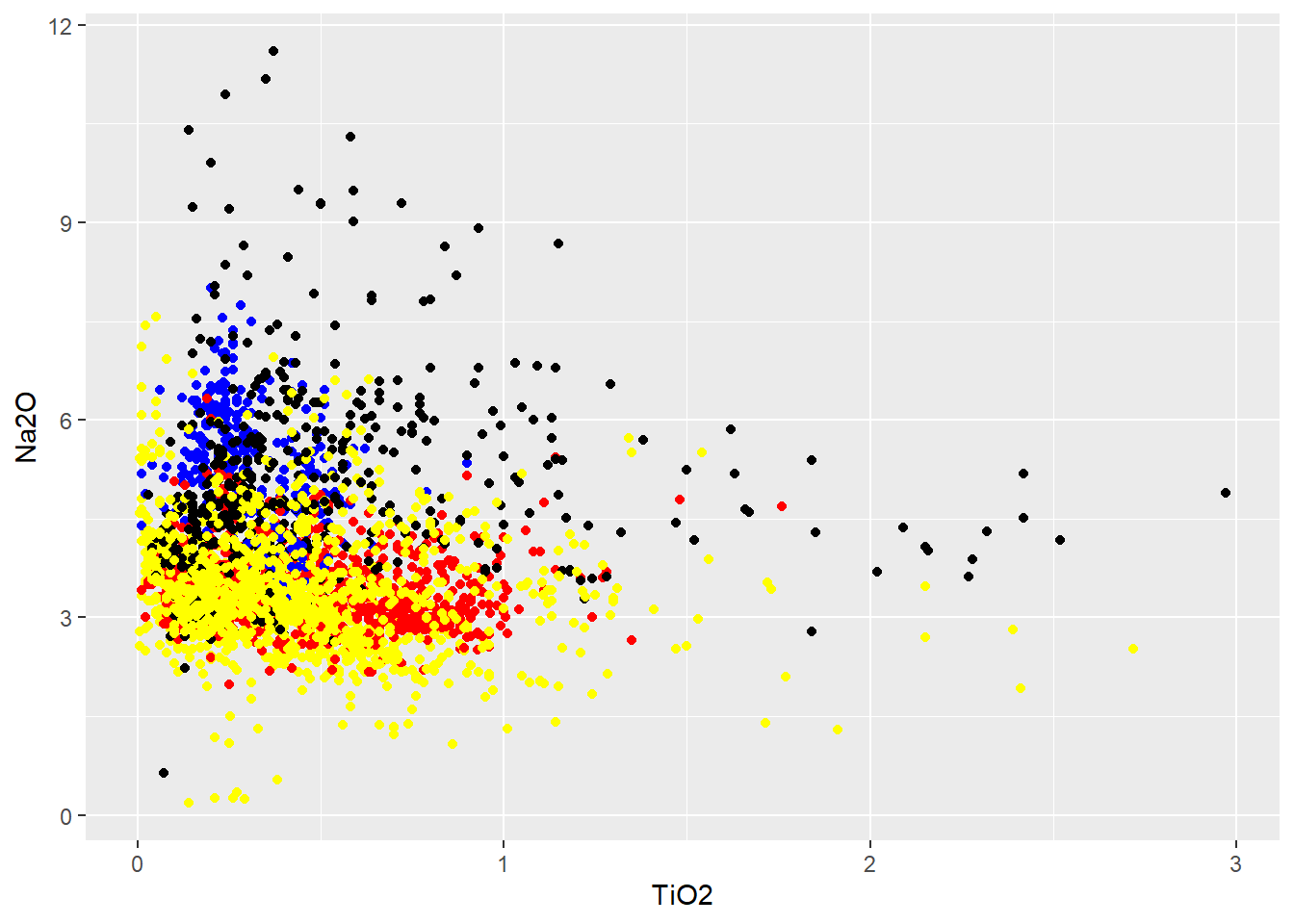
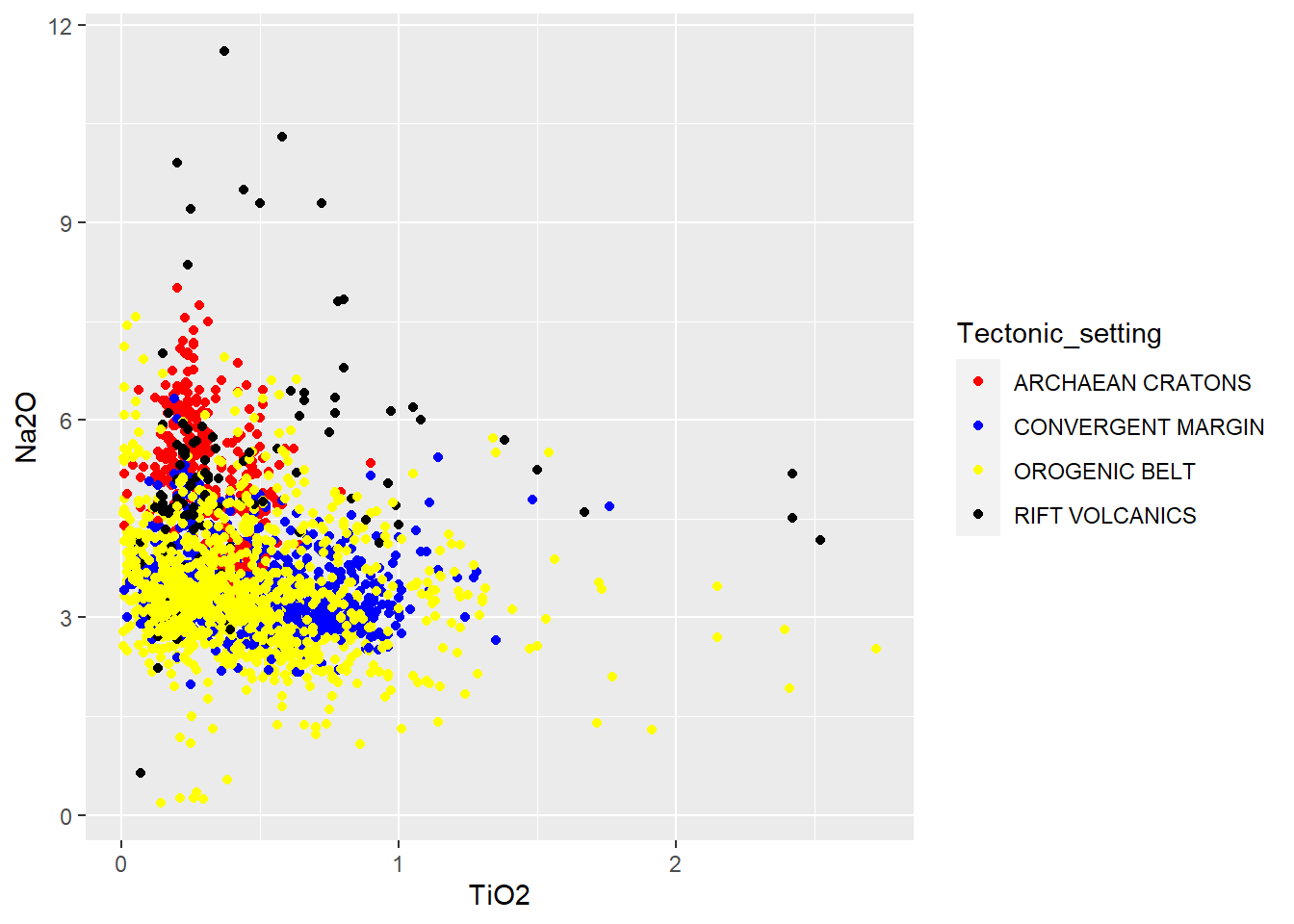
Vetor
cols<-c("red","blue","yellow")
BD %>%
filter(Tectonic_setting %in% c("CONVERGENT MARGIN", "ARCHAEAN CRATONS", "OROGENIC BELT") ) %>%
ggplot(aes(x=TiO2,y=Na2O,color=Tectonic_setting))+
geom_point()+
scale_color_manual(values=cols) 
Manuais
BD %>%
filter(Tectonic_setting %in% c("CONVERGENT MARGIN", "ARCHAEAN CRATONS", "OROGENIC BELT","RIFT VOLCANICS") ) %>%
ggplot(aes(x=TiO2,y=Na2O,color=Tectonic_setting))+
geom_point()+
scale_color_manual(values=c("red","blue","yellow","black"))